Modeling Changes in Microbiome: Focused analysis of specific genera
Last updated: 2020-06-17
Checks: 6 1
Knit directory: Fiber_Intervention_Study/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a16e6ef. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/.Rhistory
Ignored: reference-papers/Dietary_Variables.xlsx
Ignored: reference-papers/Johnson_2019.pdf
Ignored: renv/library/
Ignored: renv/staging/
Untracked files:
Untracked: analysis/glme_microbiome_genus_subset.Rmd
Untracked: analysis/glme_microbiome_phylum.Rmd
Untracked: data/analysis-data/DataDictionary_TOTALS_2018Record.xls
Untracked: fig/figure5_glmm_genus.pdf
Untracked: fig/figure5_glmm_phylum.pdf
Untracked: fig/figure5_glmm_phylum.png
Untracked: tab/results_glmm_microbiome_genus.csv
Untracked: tab/results_glmm_microbiome_phylum.csv
Untracked: tab/tables_results_2020-06-08.zip
Untracked: tab/tables_results_2020-06-08/
Unstaged changes:
Modified: analysis/data_processing.Rmd
Deleted: analysis/glme_microbiome.Rmd
Modified: analysis/index.Rmd
Modified: analysis/microbiome_diet_trends.Rmd
Modified: code/get_cleaned_data.R
Modified: code/microbiome_statistics_and_functions.R
Modified: fig/figure2.pdf
Modified: fig/figure3.pdf
Modified: fig/figure4.pdf
Modified: fig/figure4_final_microbiome_diet_variables_over_time.pdf
Modified: fig/figure4_legend.pdf
Modified: fig/figure5_glmm.pdf
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
This page contains the investigation of the changes over time:
Lachnospira, Akkermannsia, Bifidobacterium, Lactobacillus, Ruminococcus, Roseburia, Clostridium, Faecalibacterium, and Dorea
Prevotella/Bacteroides
Data prep
This is only to recode some variables to ease interpretation.
microbiome_data$meta.dat <- microbiome_data$meta.dat %>%
mutate(intB = ifelse(Intervention=="B", 1,0),
time = as.numeric(Week) - 1,
female = ifelse(Gender == "F", 1, 0),
hispanic = ifelse(Ethnicity %in% c("White", "Asian", "Native America"), 1, 0))
# get specific genera
N <- nrow(microbiome_data$meta.dat)
ids <- microbiome_data$meta.dat$ID
dat <- data.frame(microbiome_data$abund.list[["Genus"]])
# NOTE: Special coding for stripping weird characters from bacteria names
j <- length(rownames(dat))
i <- 1
for(i in 1:j){
while( substring(rownames(dat[i,]), 1, 1) == "_"){
if(i == 1){
row.names(dat) <- c(substring(rownames(dat[i,]), 2),rownames(dat)[2:j])
}
if(i > 1 & i < j){
row.names(dat) <- c(rownames(dat[1:(i-1),]),
substring(rownames(dat[i,]), 2),
rownames(dat)[(i+1):j])
}
if(i == j){
row.names(dat) <- c(rownames(dat[1:(j-1),]),substring(rownames(dat[j,]), 2))
}
} # End while loop
} # End for loop
# ====================== #
num.bact <- nrow(dat)
dat <- t(dat[1:num.bact,1:N])
# subset to specific bacteria
bactNames <- c("Akkermansia", "Bacteroides", "Bifidobacterium", "Clostridium_sensu_stricto_1", "Dorea", "Faecalibacterium", "Lachnospira", "Lactobacillus", "Prevotella", "Roseburia", "Ruminococcus_1")
backNames <- bactNames[order(bactNames)]
dat <- dat[, bactNames]
dat <- apply(dat, 2, function(x){log(x+1)})
k <- ncol(microbiome_data$meta.dat) # number of original variables
dat <- data.frame(cbind(microbiome_data$meta.dat, dat[ids,]))Note: for Clostridium and Ruminococcus we had to select one out of multiple potential reads due to uncertainty in the algorithm. We selected the closest match for each by using the Clos. or Rum. that had the most reads. The choices we had are shown below
B<- as.matrix(microbiome_data$abund.list$Genus)
B <- t(B)
Bt <- colSums(B)
# Clos
Bt[names(Bt) %like% "Clostridium"]___Clostridium_innocuum_group __Clostridium_sensu_stricto_1
4 204 # Rum.
Bt[names(Bt) %like% "Ruminococcus"]___Ruminococcus_gauvreauii_group ___Ruminococcus_torques_group
3 600
__Ruminococcus_1 __Ruminococcus_2
3238 1298 Genus Level Microbiome and Intervention
We will build up to the effect of the intervention by investigating covariates to see if we can exclude them from model with intervention. Due to only having 11 people, we will only estimate 1 random effect per model (random intercepts). All other effects are fixed, but we will only estimate up to 2 effects. Meaning we can only include two covariates in each model at a time.
In all models, \(h(.)\) is an arbitrary link function. We selected the Poisson model for the outcomes so the link function is the log link.
For all significance test, we will use \(p < 0.05\) as the level to declare significance, but we will largely focus on magnitude of effects.
Unconditional Model
This is a random intercepts model (worst case). No covariates are included.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \end{align*}\]
bactNames <- c("Akkermansia", "Bacteroides", "Bifidobacterium", "Clostridium_sensu_stricto_1", "Dorea", "Faecalibacterium", "Lachnospira", "Lactobacillus", "Prevotella", "Roseburia", "Ruminococcus_1")
genus.fit0 <- glmm_microbiome(mydata=microbiome_data, model.number=0,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + (1|SubjectID)")
#########################################
#########################################
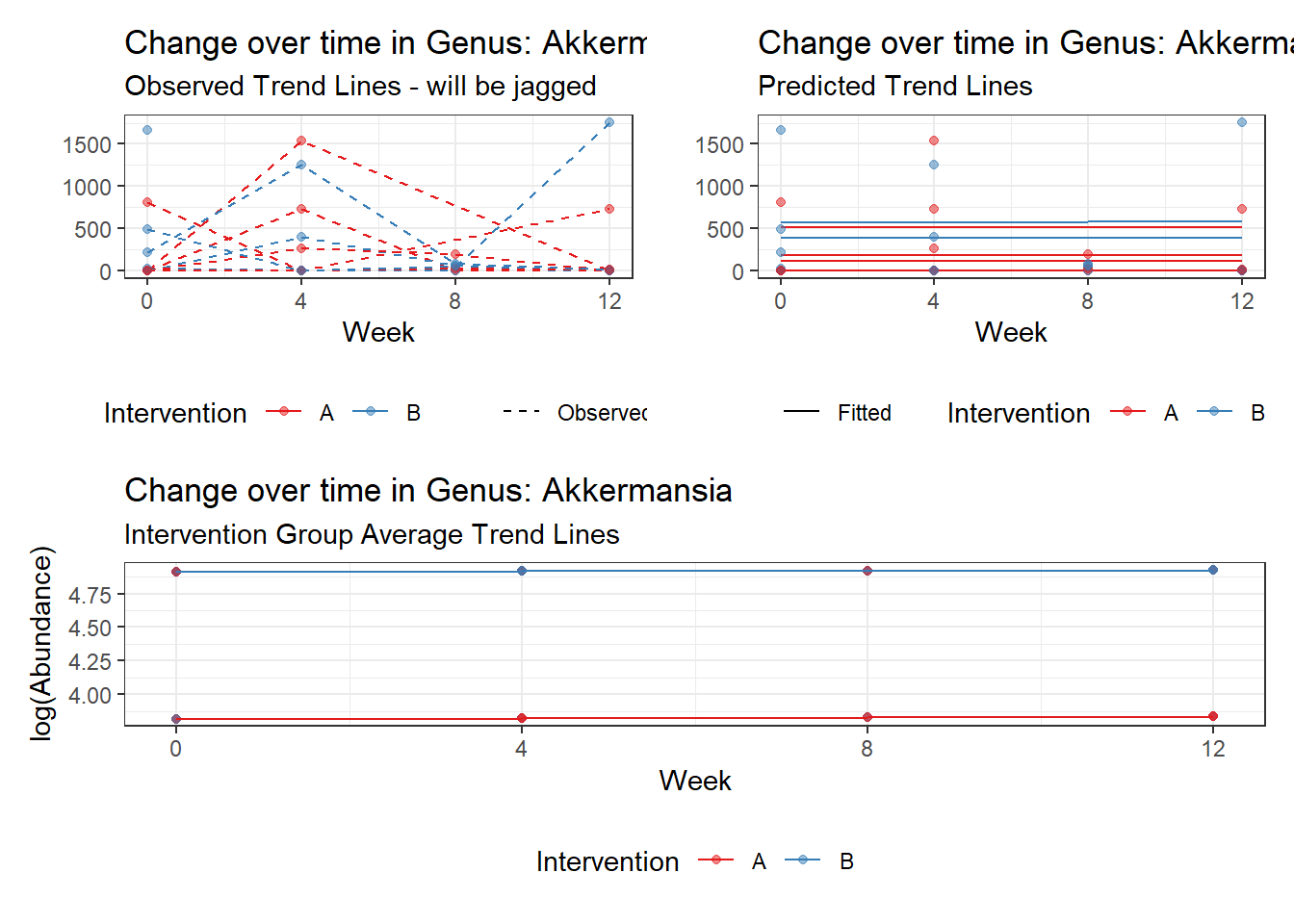
Model: Akkermansia ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.8607519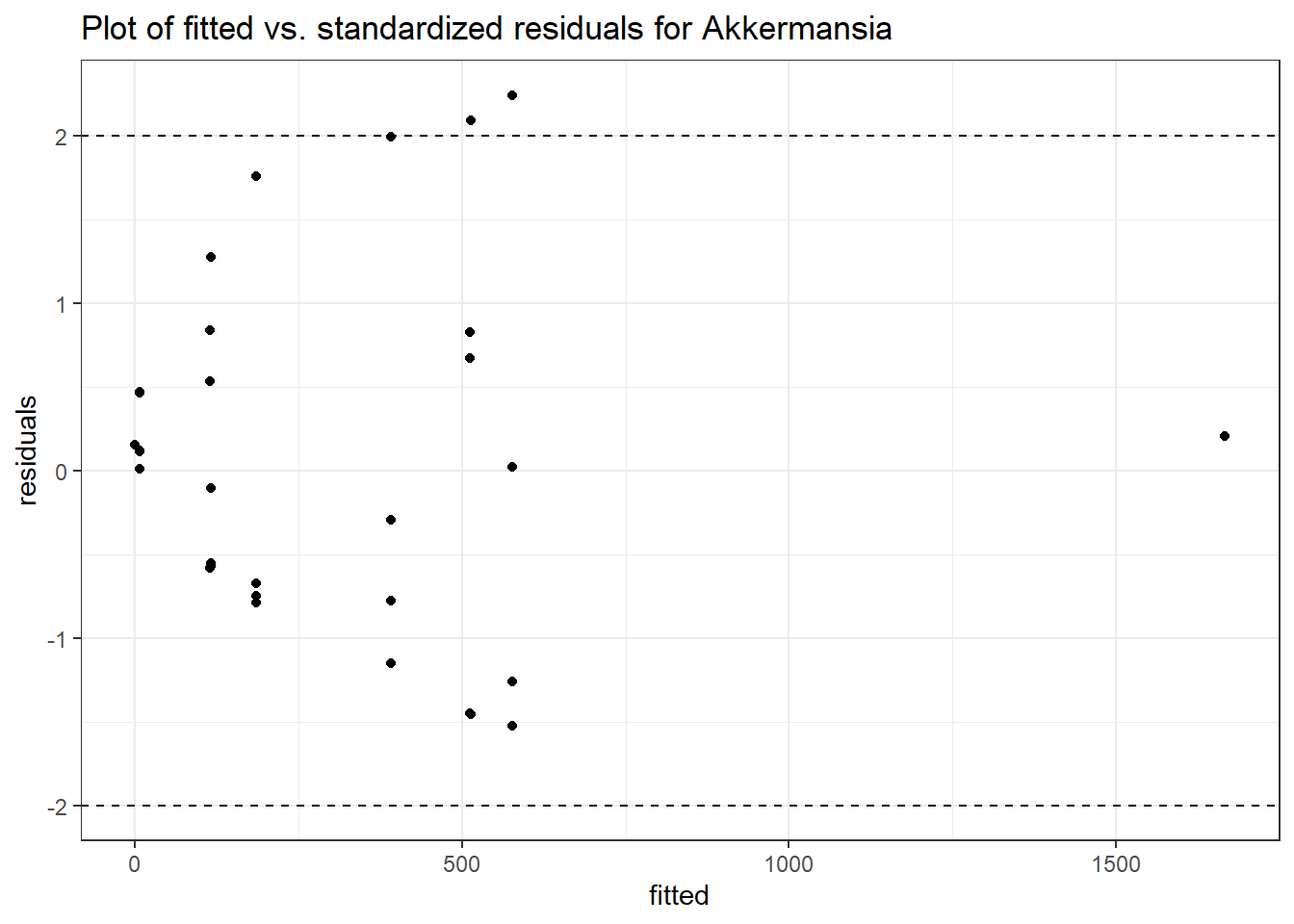
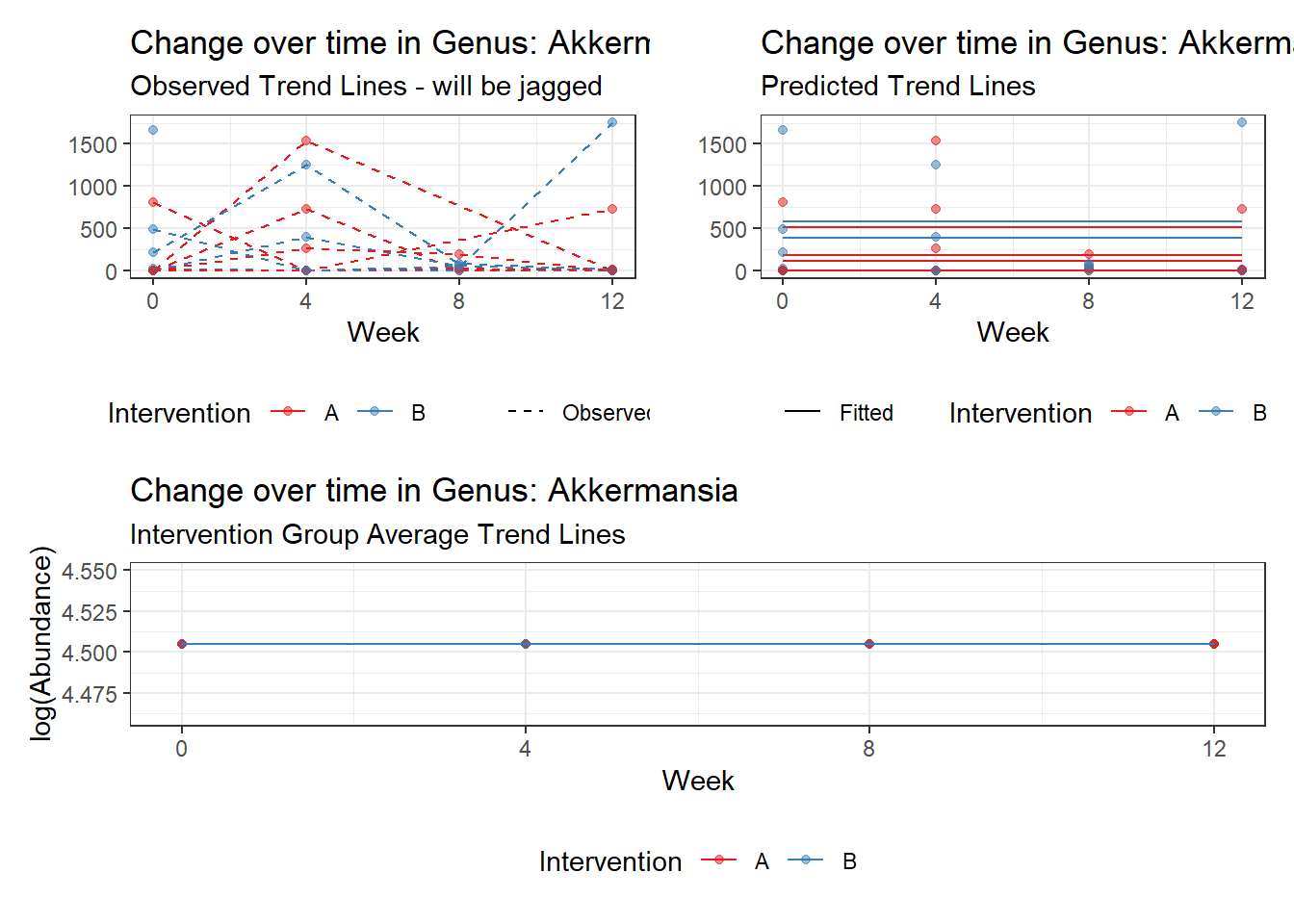
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 4.50476 0.75627 5.95654 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4862
Significance test not available for random effects
#########################################
#########################################
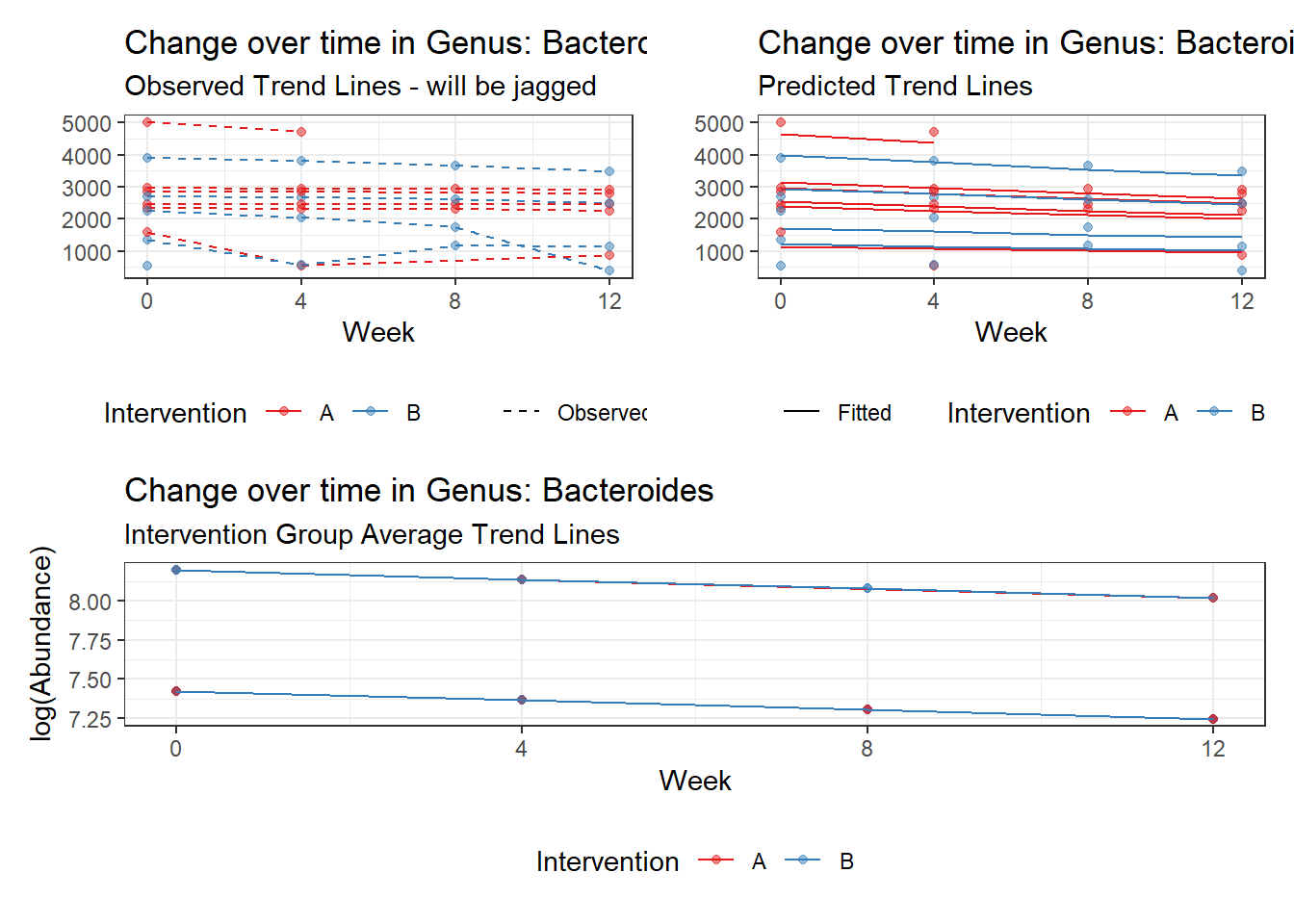
Model: Bacteroides ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.276689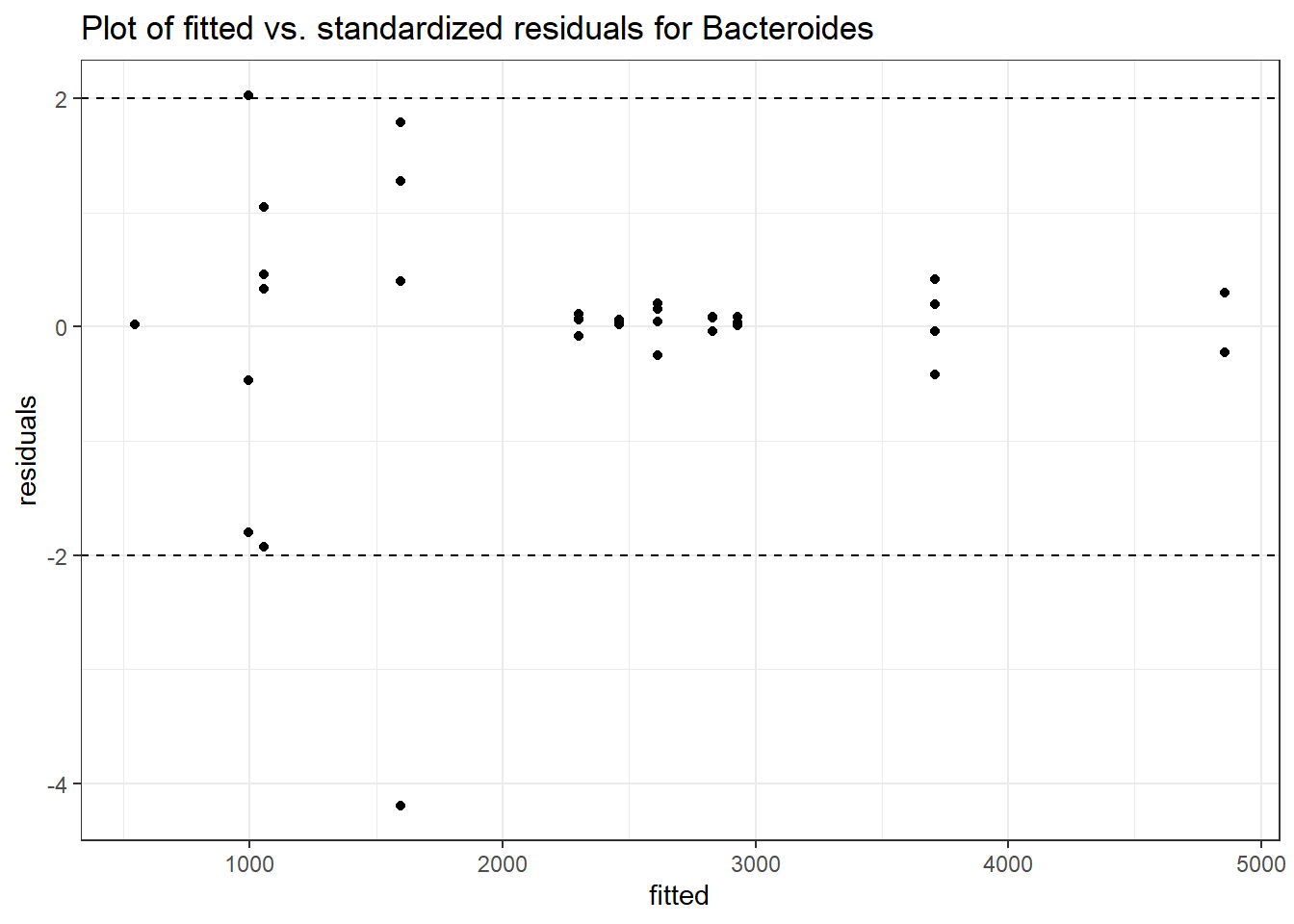
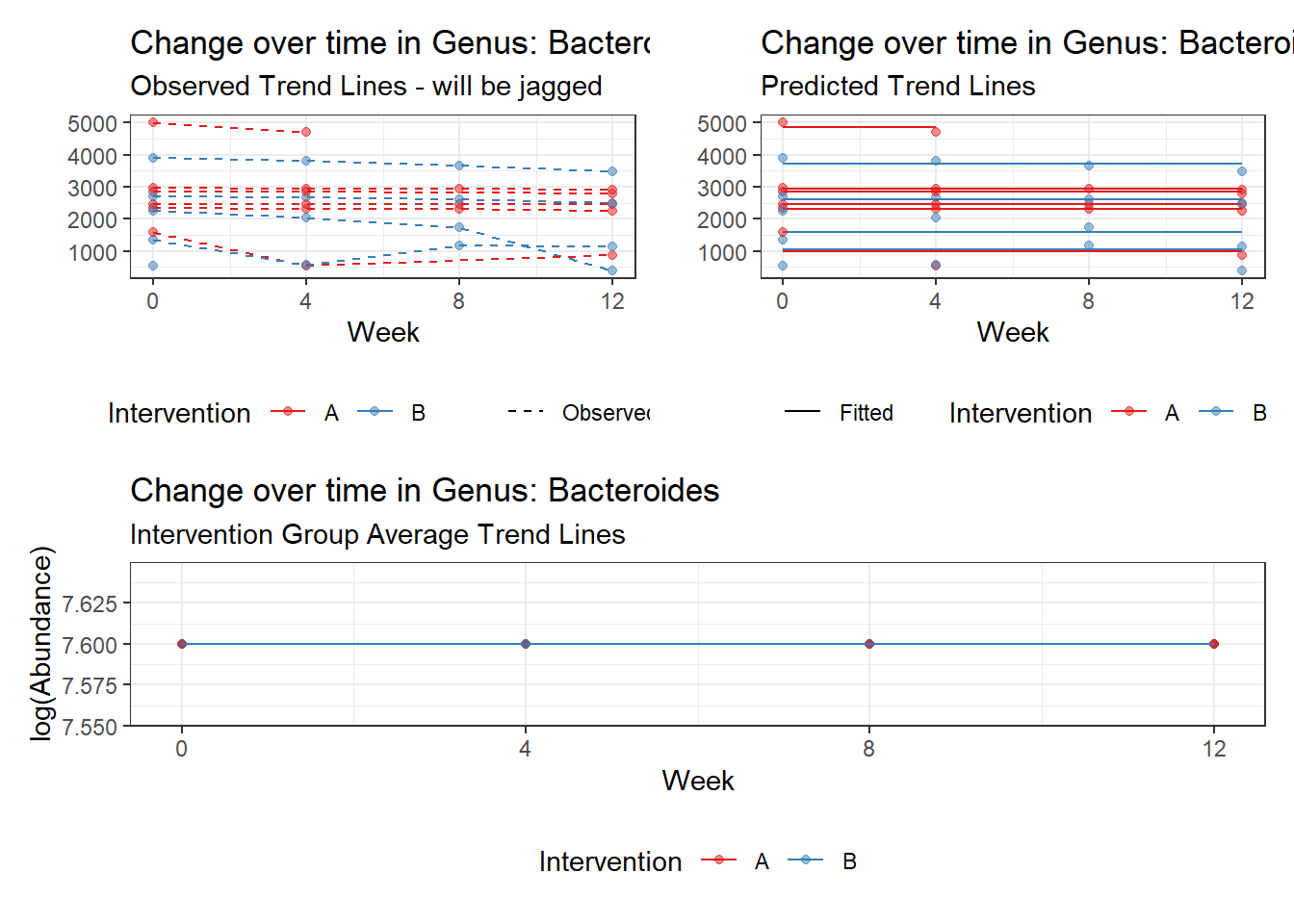
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.59968 0.18655 40.7371 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.61849
Significance test not available for random effects
#########################################
#########################################
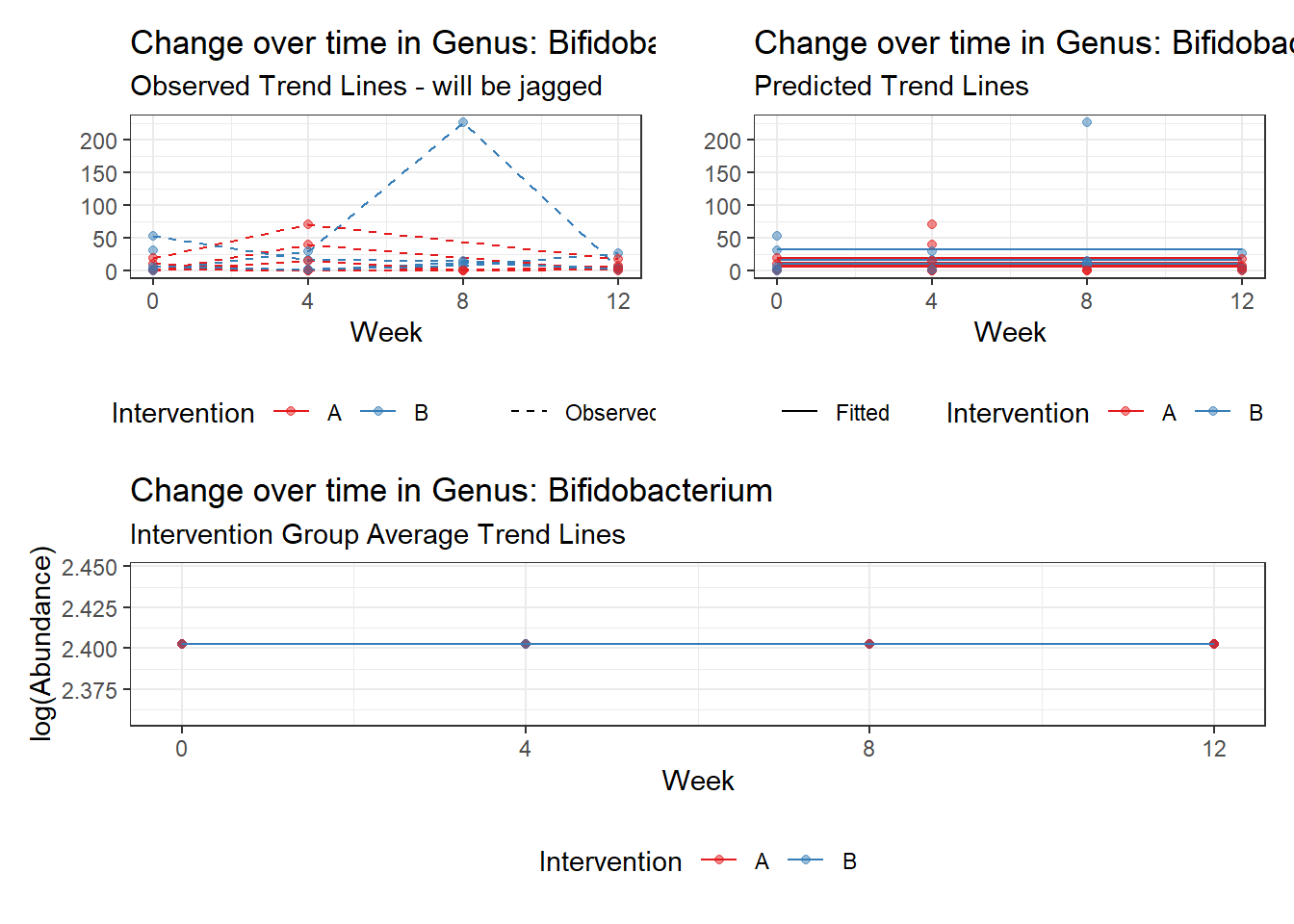
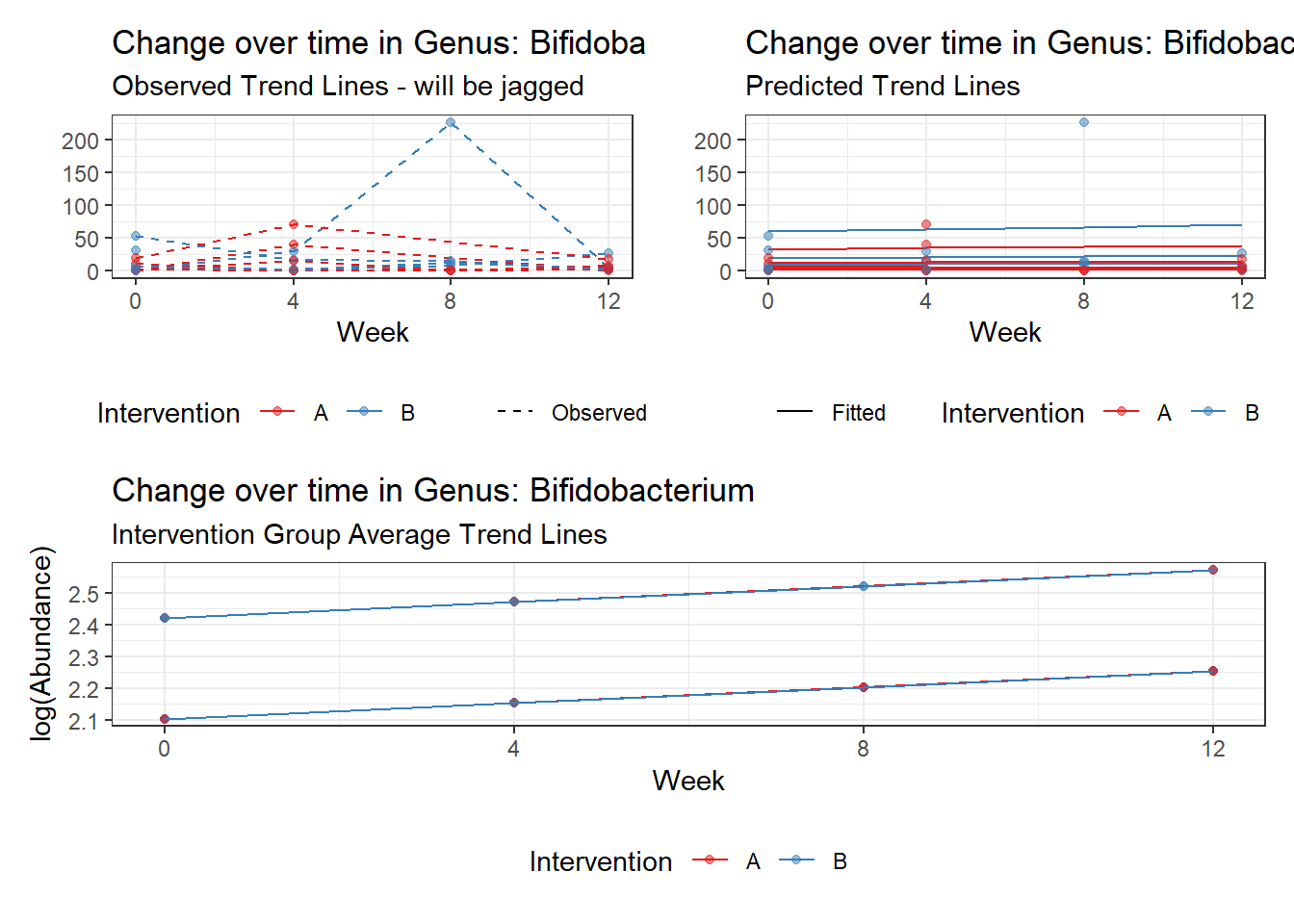
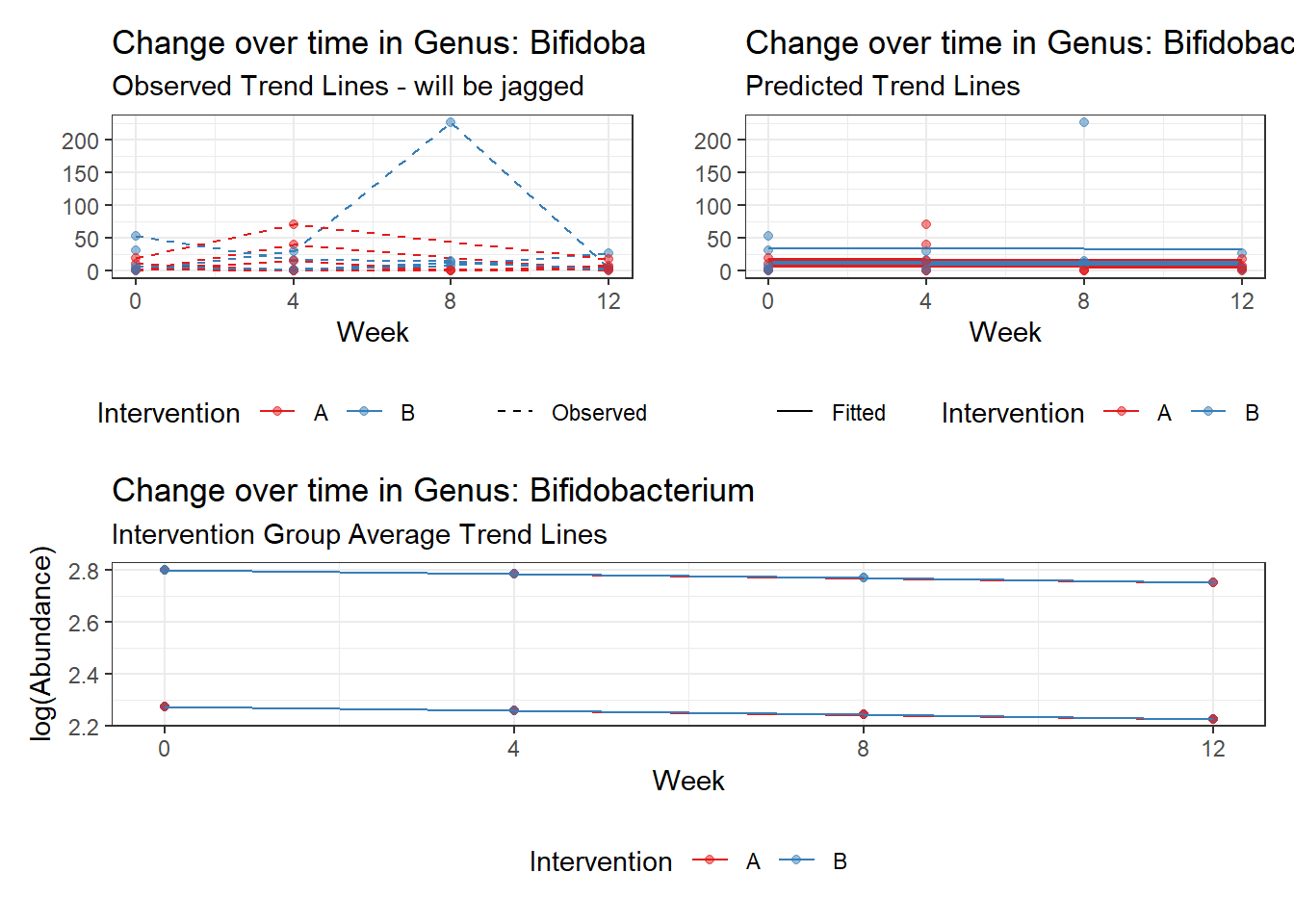
Model: Bifidobacterium ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.5660048
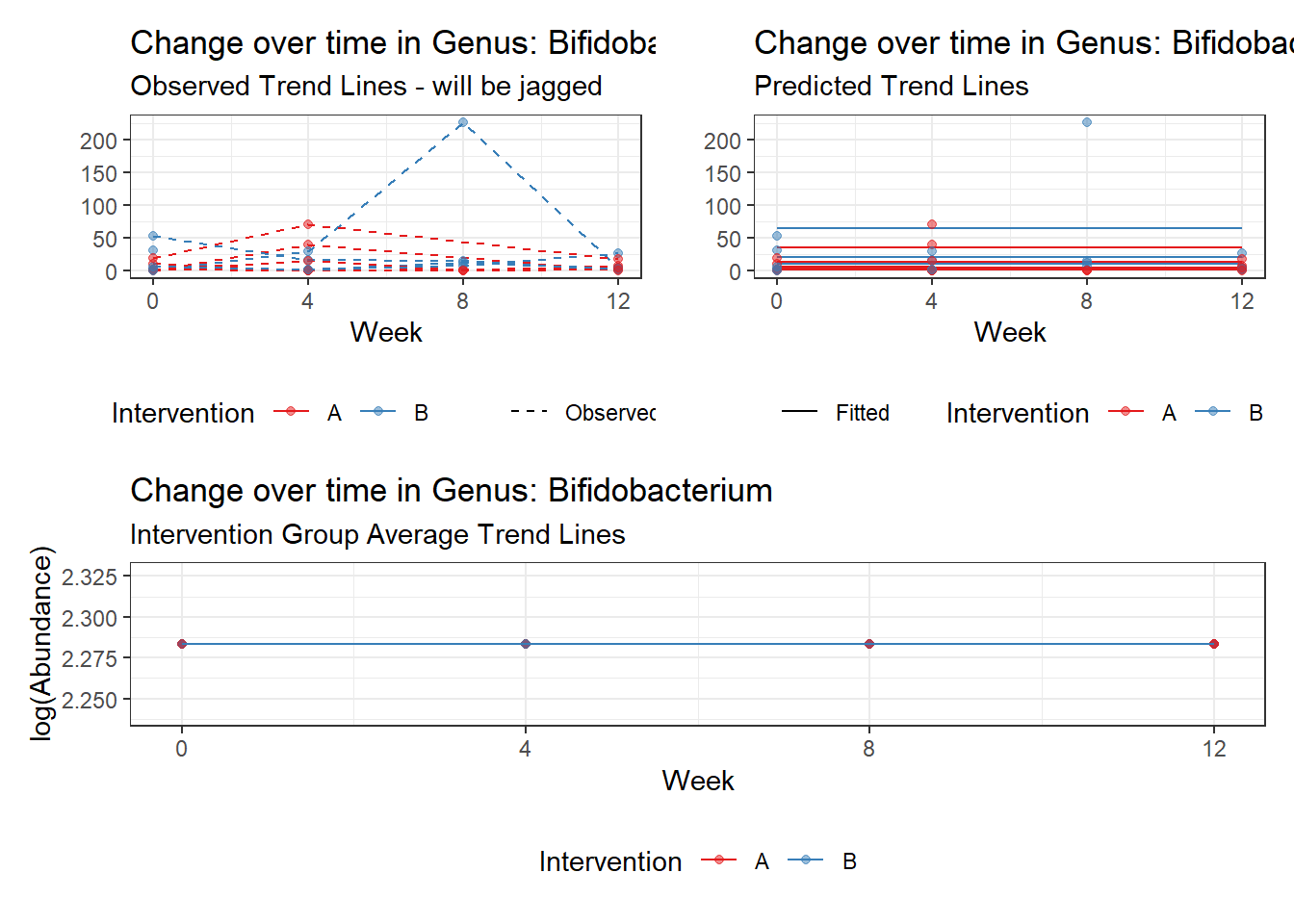
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.28319 0.35091 6.50647 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.142
Significance test not available for random effects
#########################################
#########################################
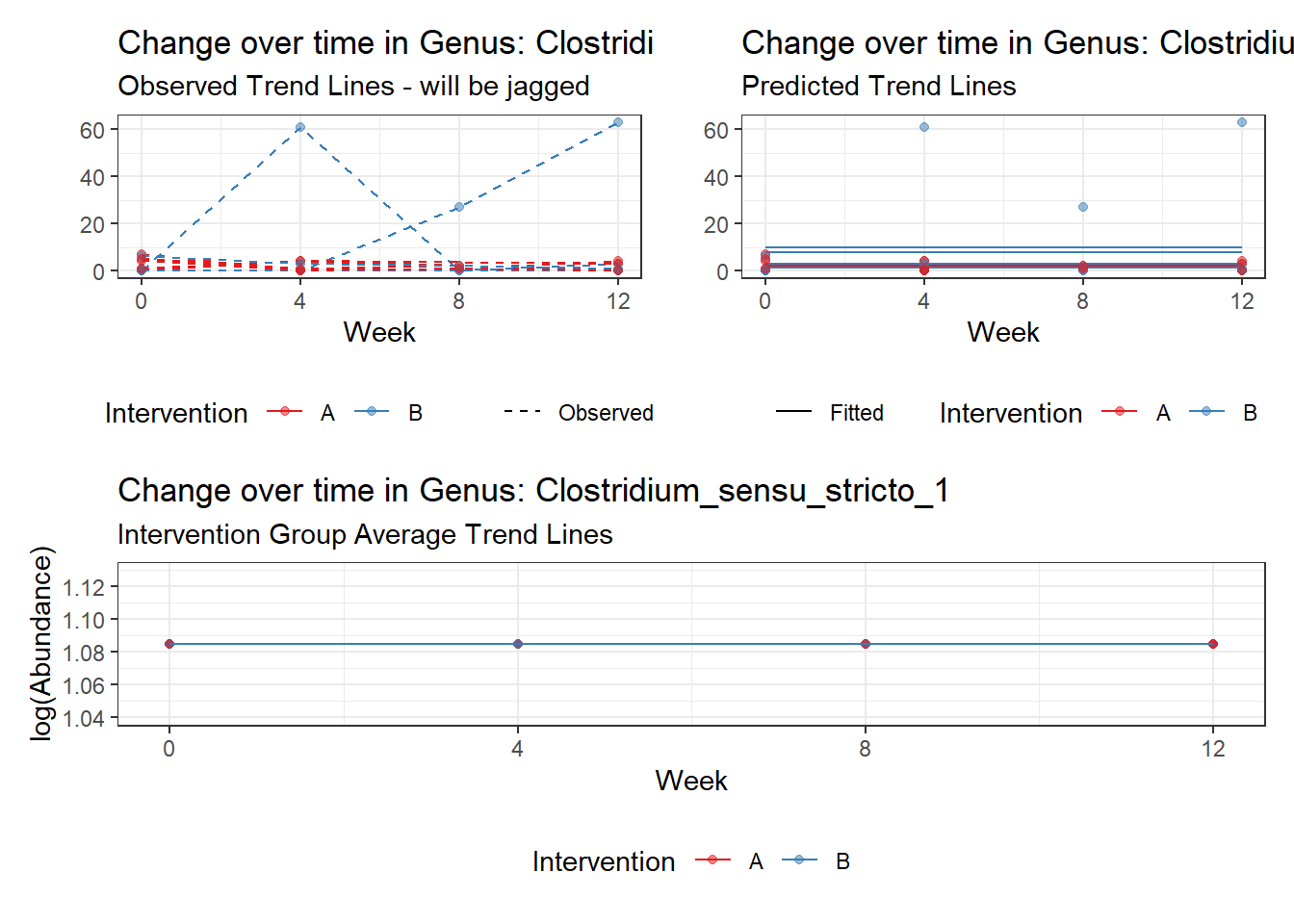
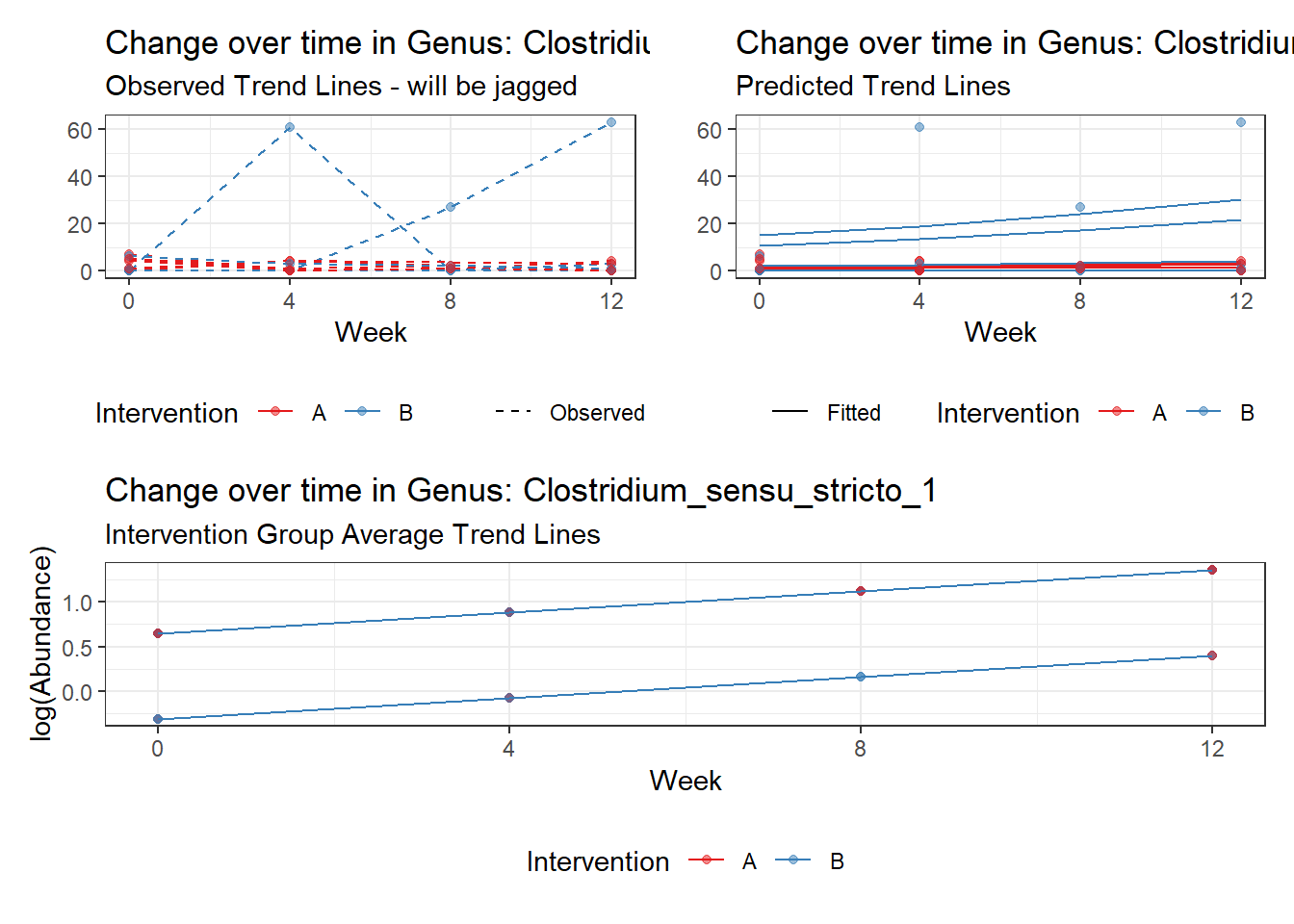
Model: Clostridium_sensu_stricto_1 ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.668022
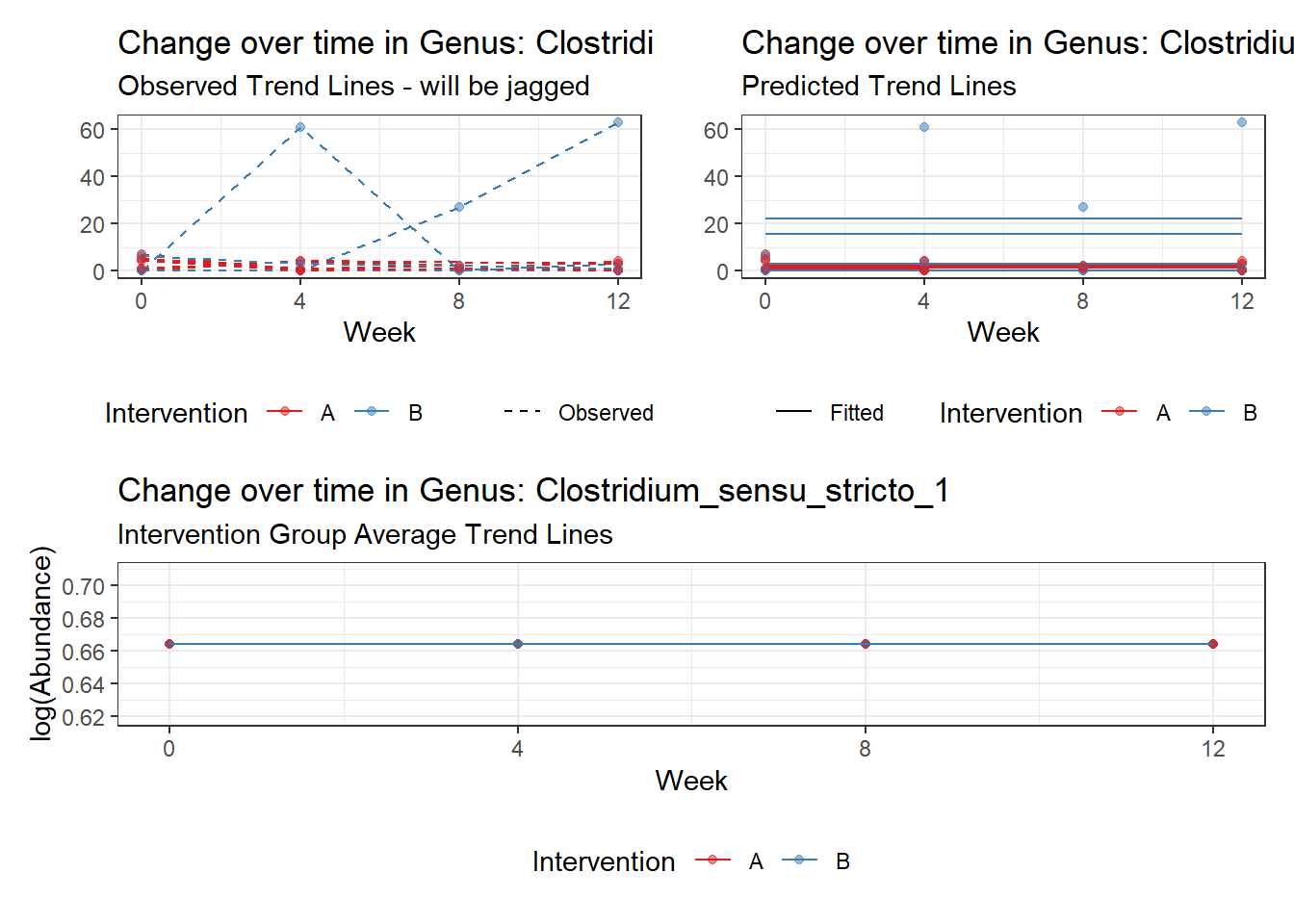
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.66389 0.46636 1.42357 0.15457
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4185
Significance test not available for random effects
#########################################
#########################################
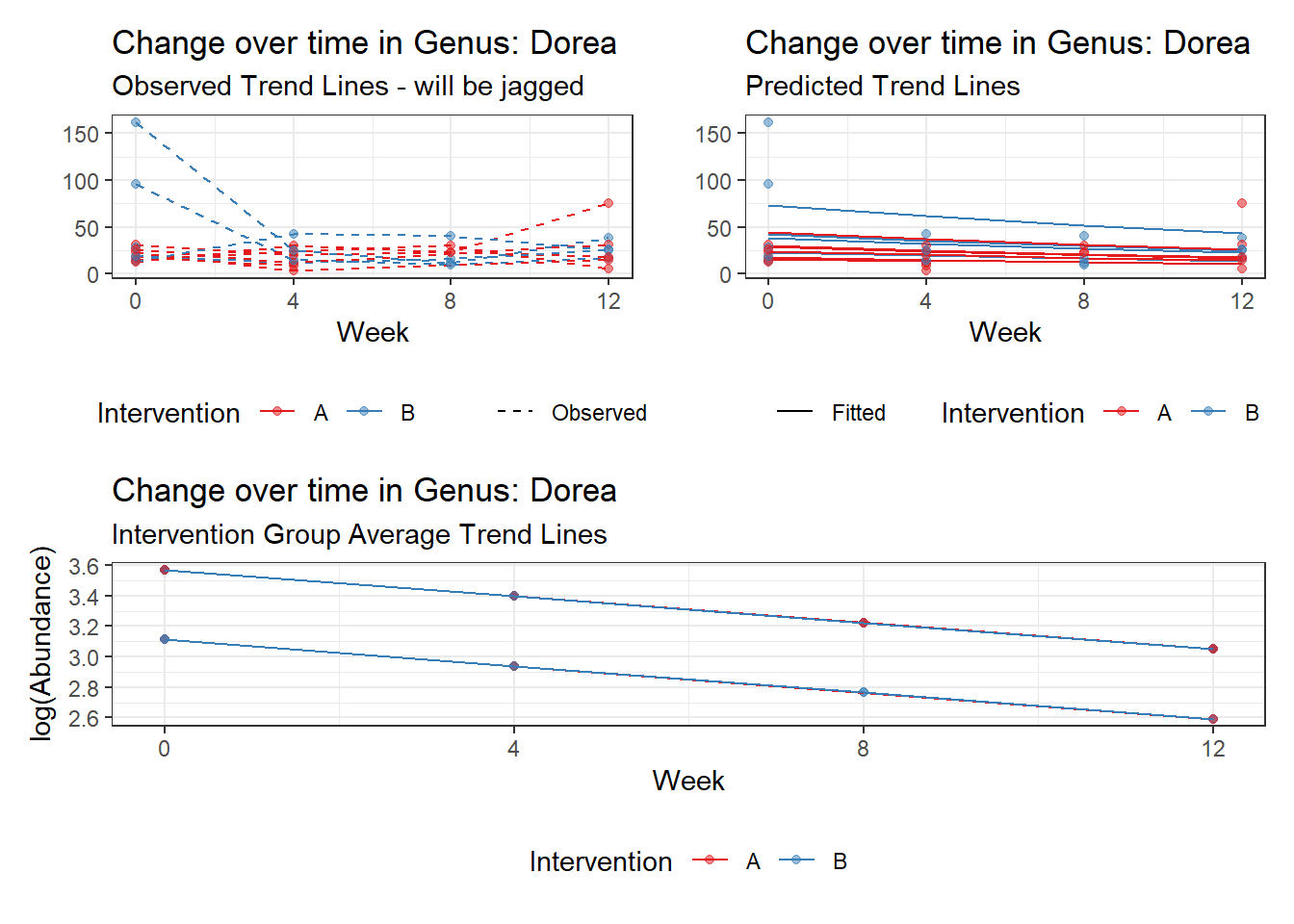
Model: Dorea ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.1456313

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.20177 0.13007 24.61638 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.41286
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.3329988
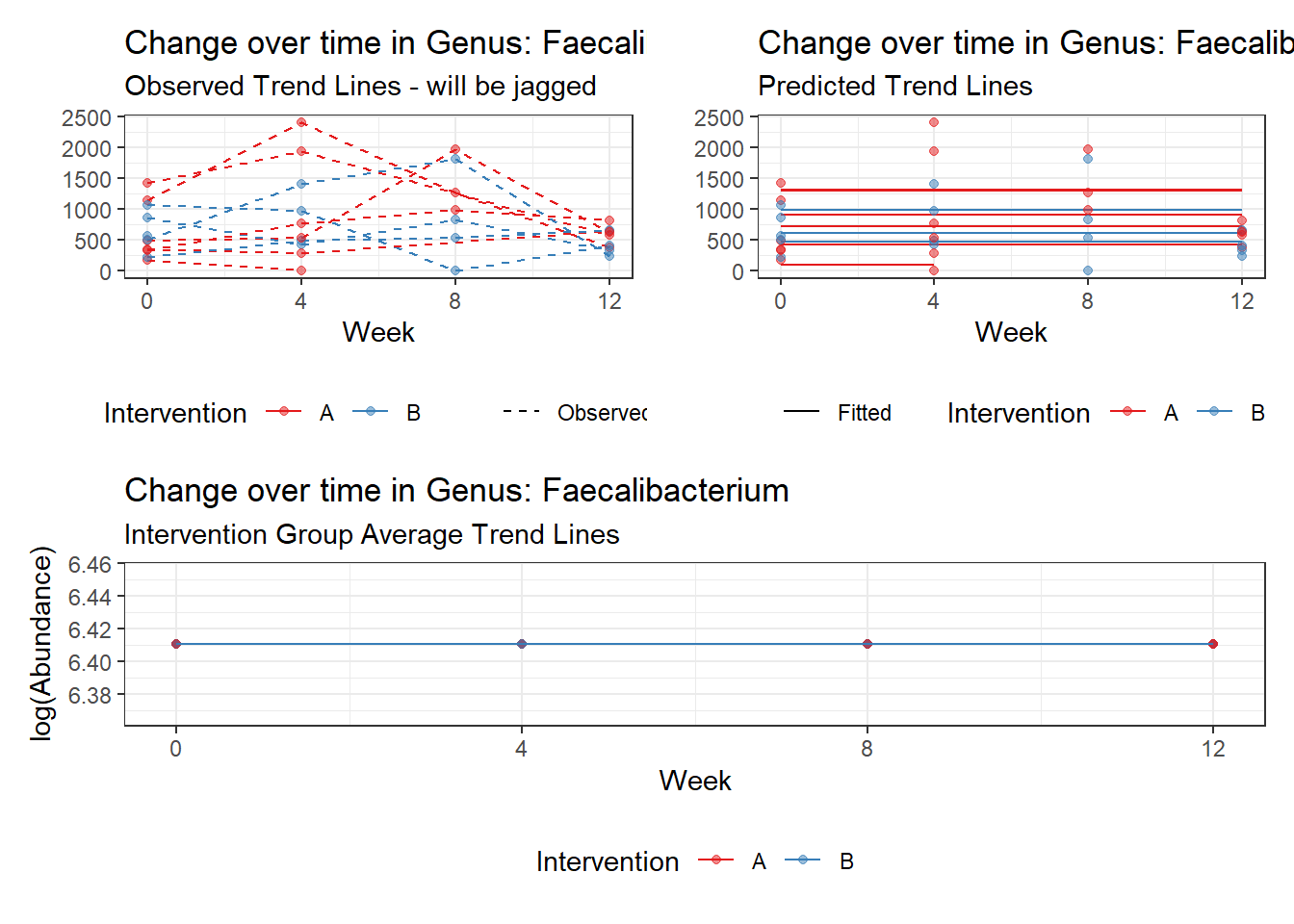
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.41054 0.21325 30.06113 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.70657
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.3651649

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 3.95517 0.23081 17.13574 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.75843
Significance test not available for random effects
#########################################
#########################################
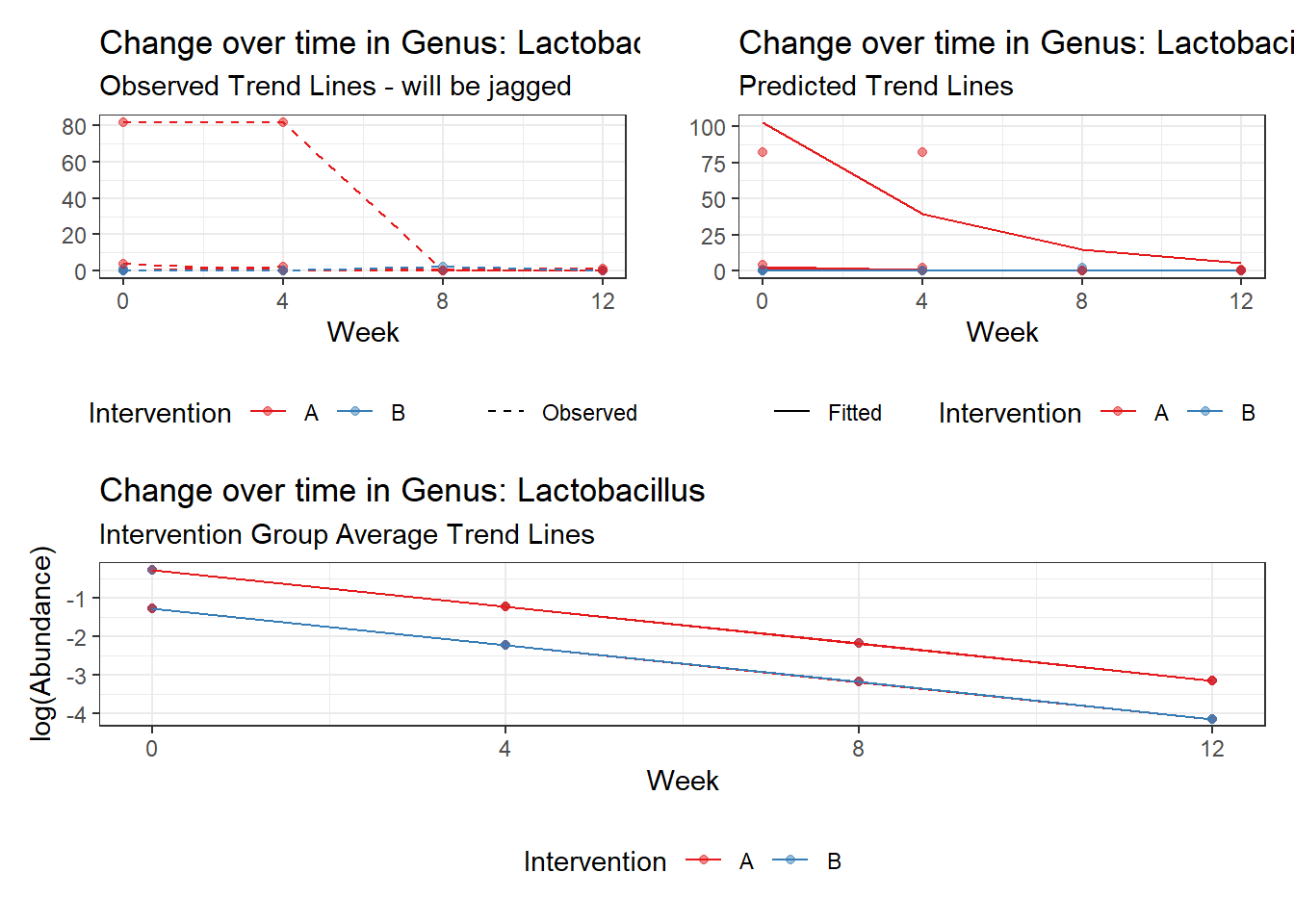
Model: Lactobacillus ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.8670383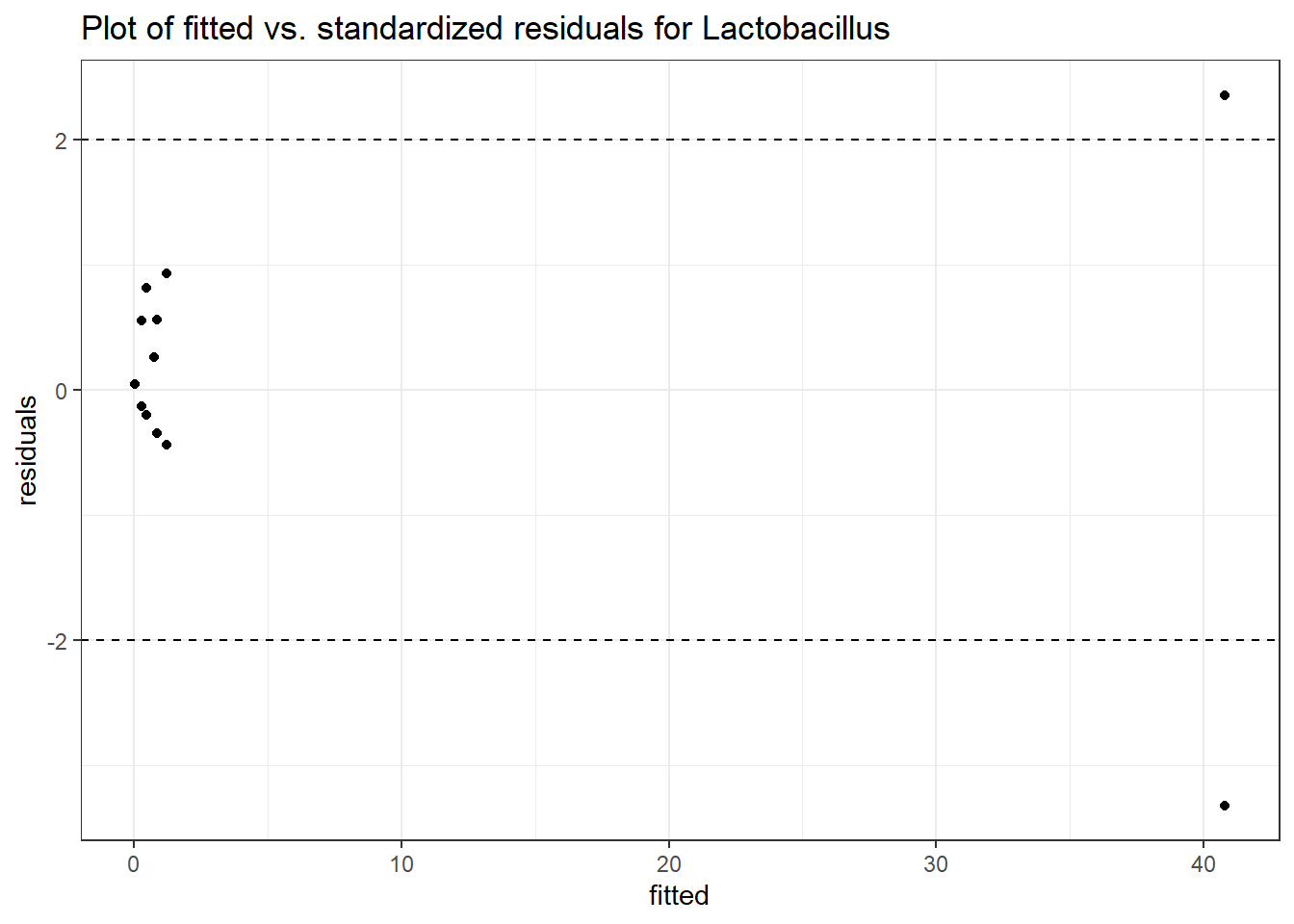

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus -1.71857 1.0382 -1.65534 0.09785
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.5536
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.7597881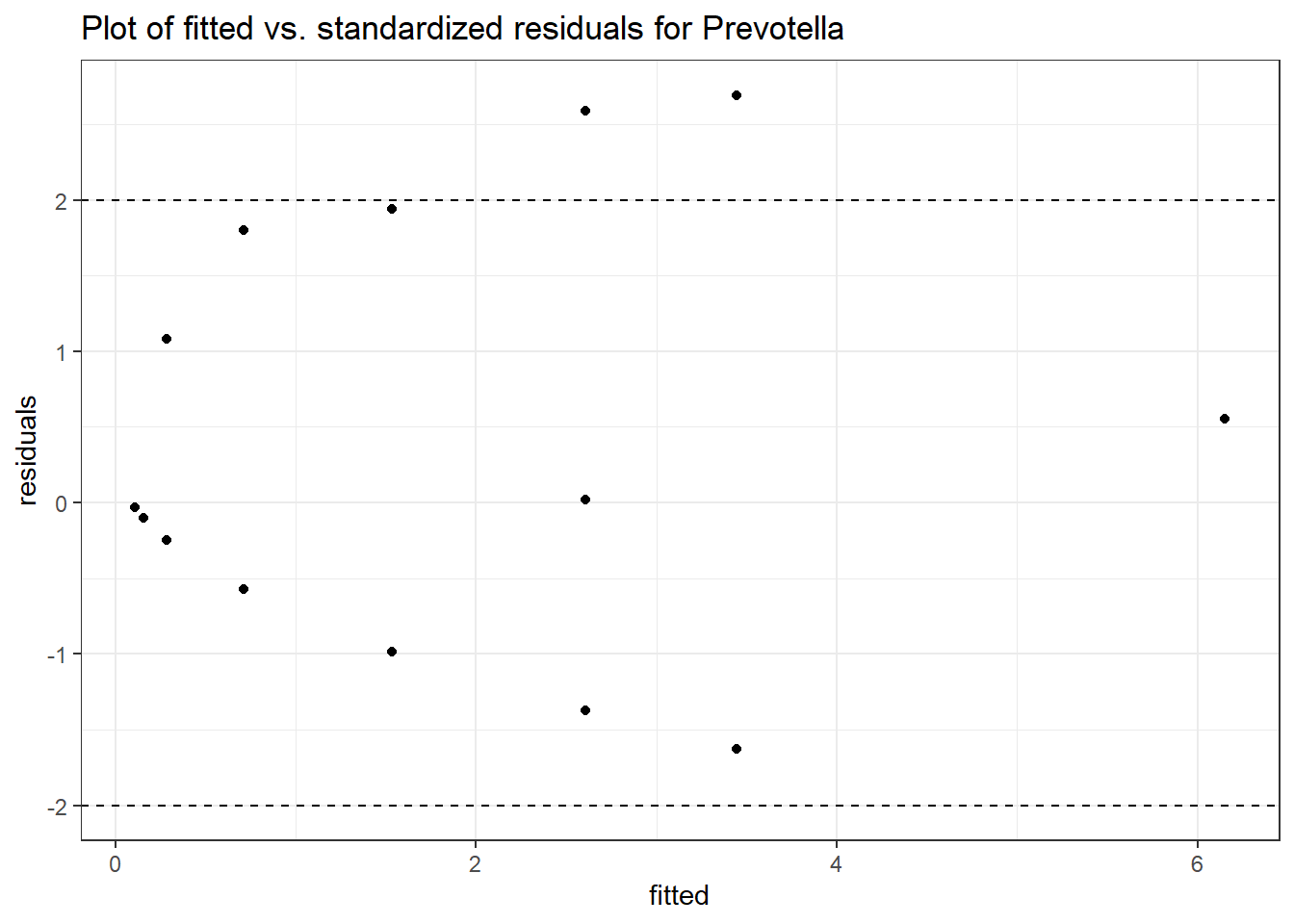
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -0.86032 0.68764 -1.25112 0.21089
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7785
Significance test not available for random effects
#########################################
#########################################
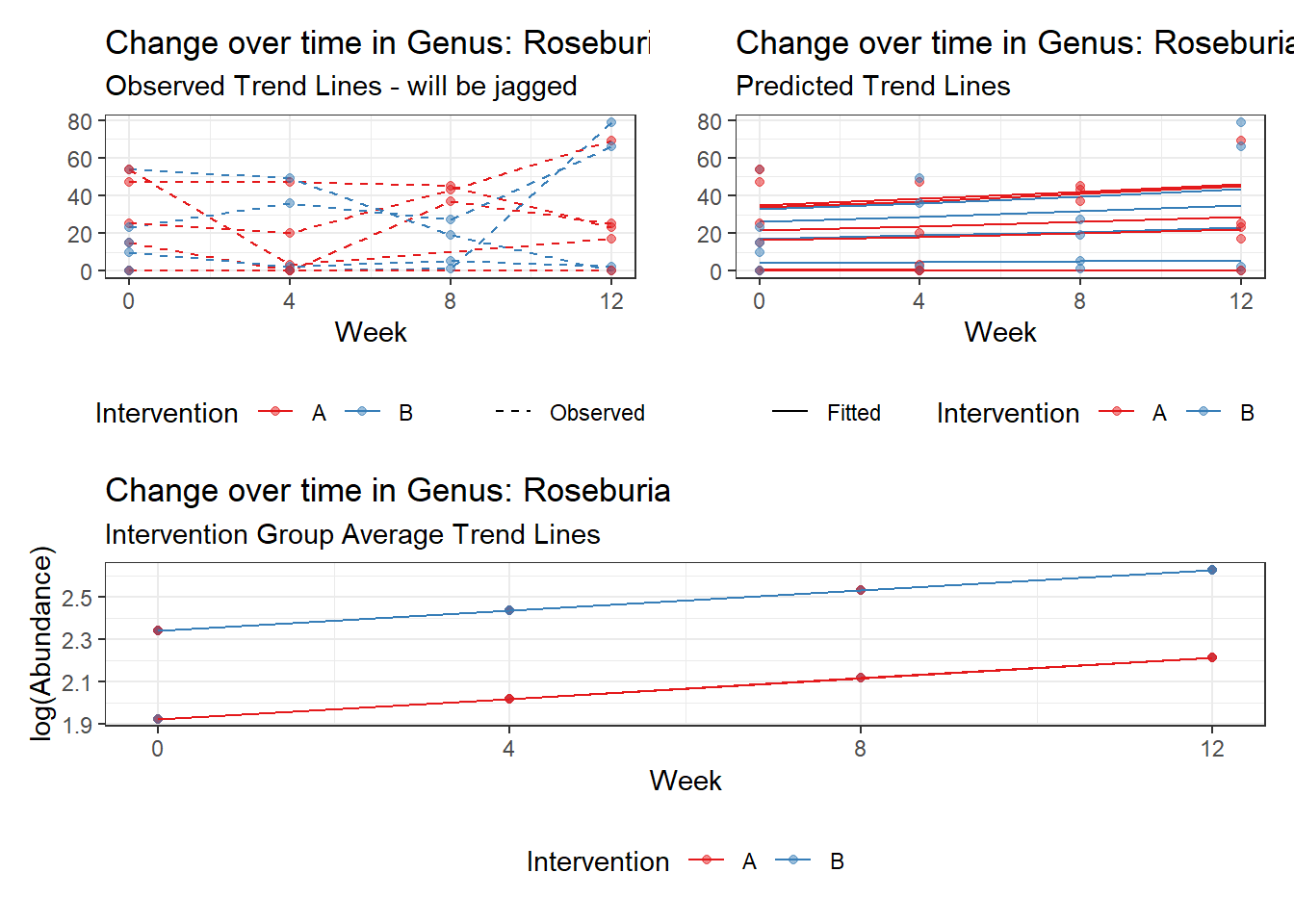
Model: Roseburia ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.7605864
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 2.30855 0.55696 4.14494 3e-05
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7824
Significance test not available for random effects
#########################################
#########################################
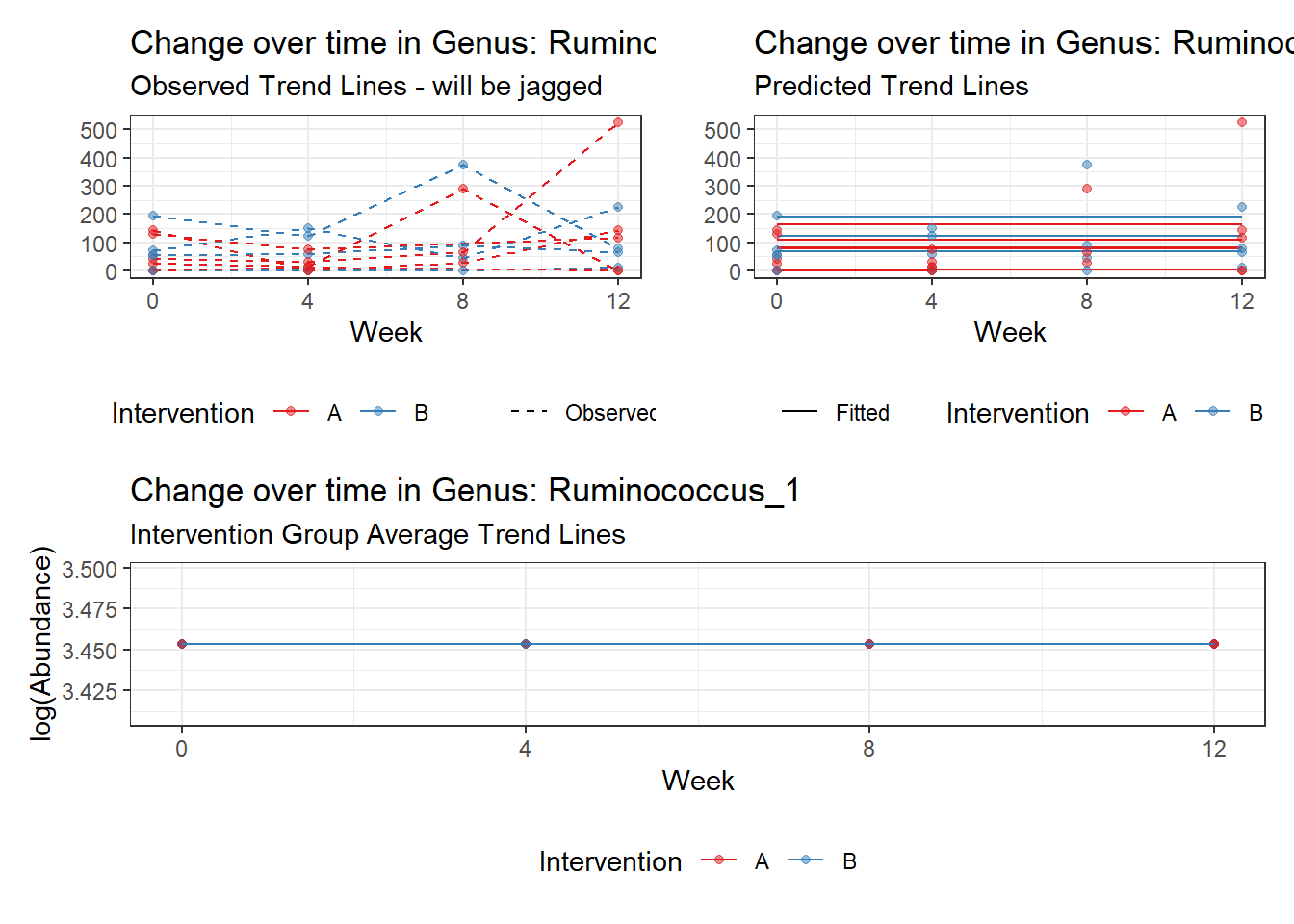
Model: Ruminococcus_1 ~ 1 + (1 | SubjectID)
<environment: 0x000000003e2d3e48>
Link: poisson
Intraclass Correlation (ICC): 0.7992502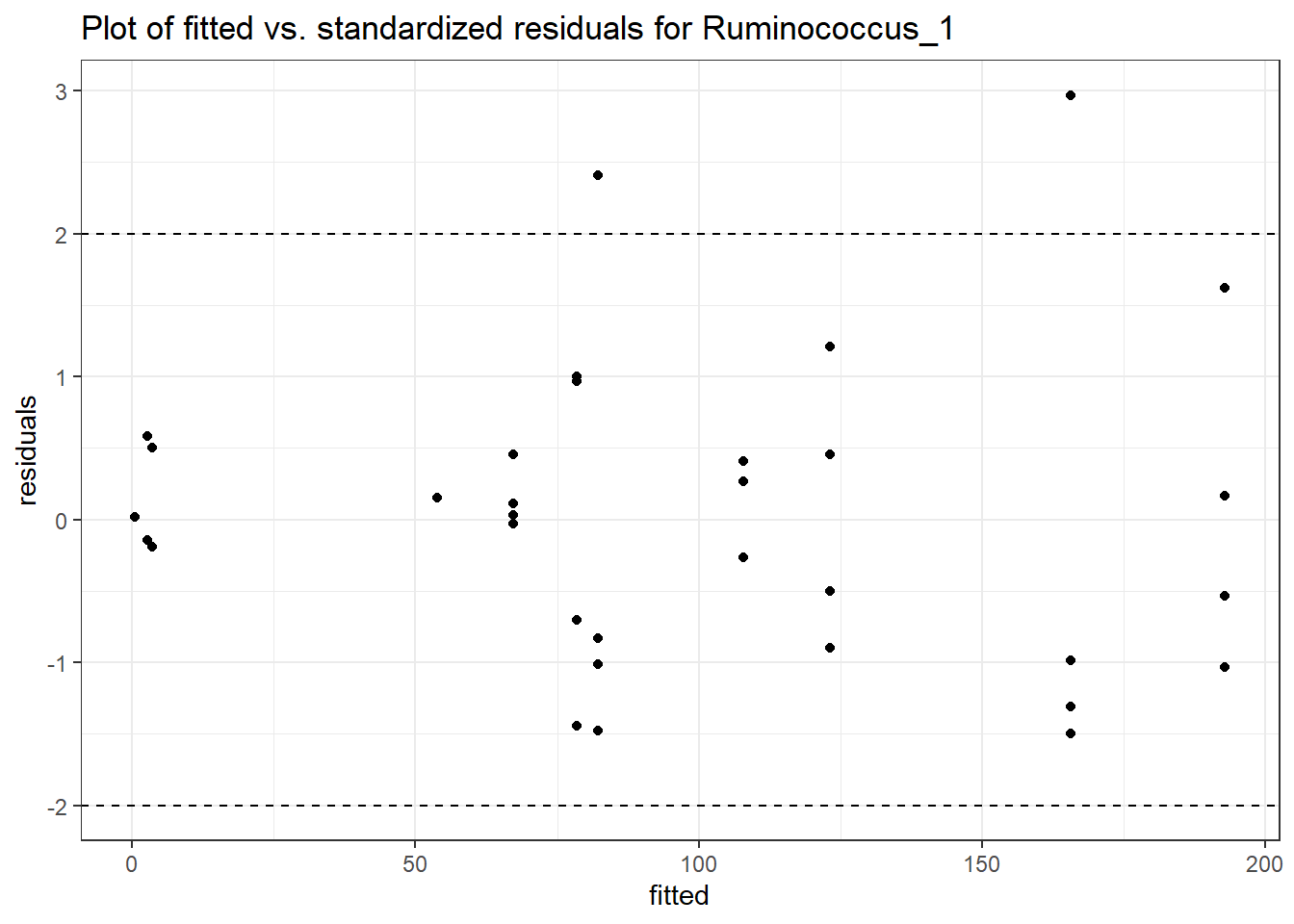
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 3.45334 0.60983 5.66276 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.9953
Significance test not available for random effectsgenus.fit0b <- glmm_microbiome(mydata=microbiome_data, model.number=0,
taxa.level="Genus", taxa.subset = backNames,
link="negbinom",
model="1 + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced
Intraclass Correlation (ICC): 0.8611513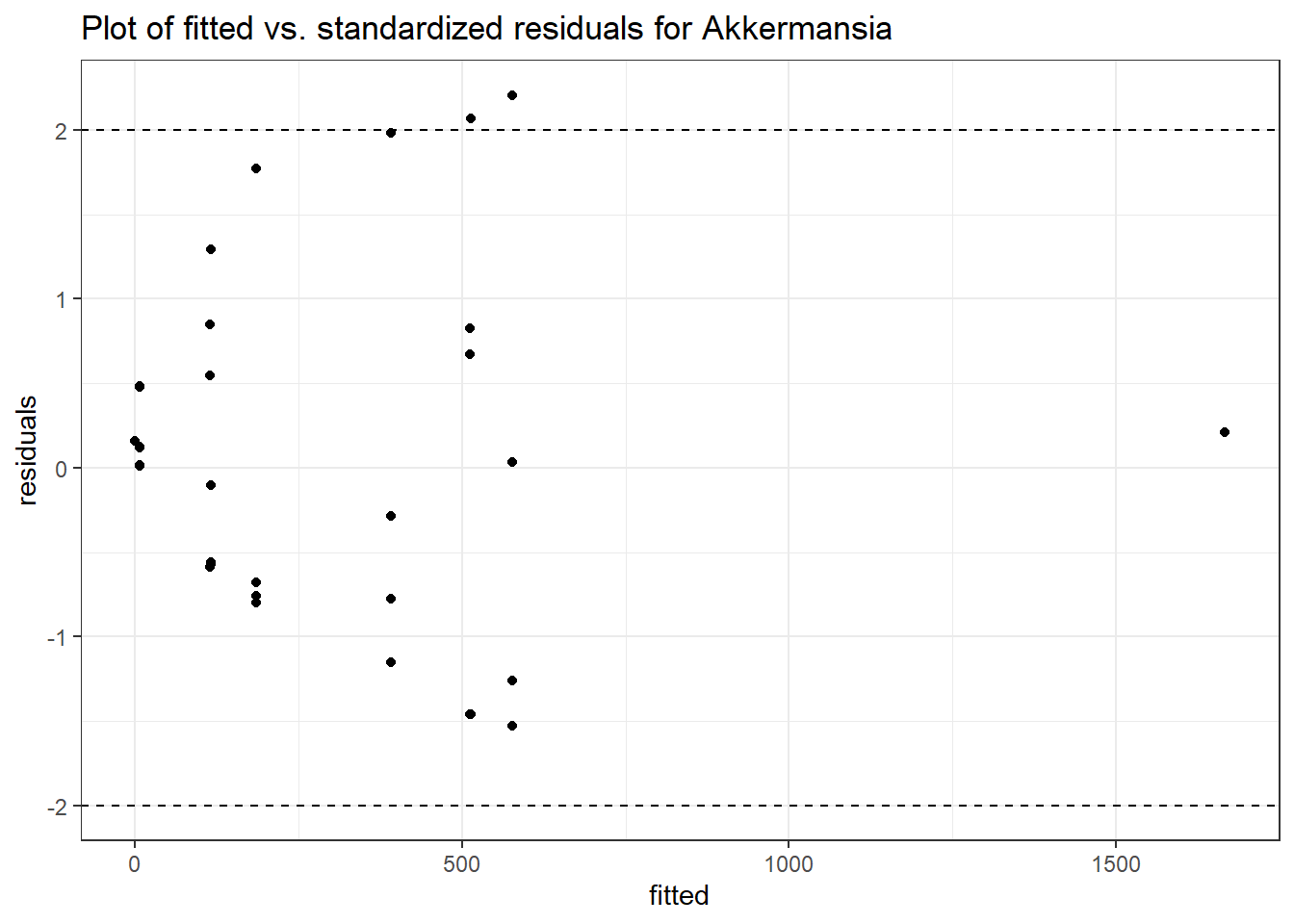

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 4.50251 0.75647 5.95201 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4904
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinom
Intraclass Correlation (ICC): 0.2254147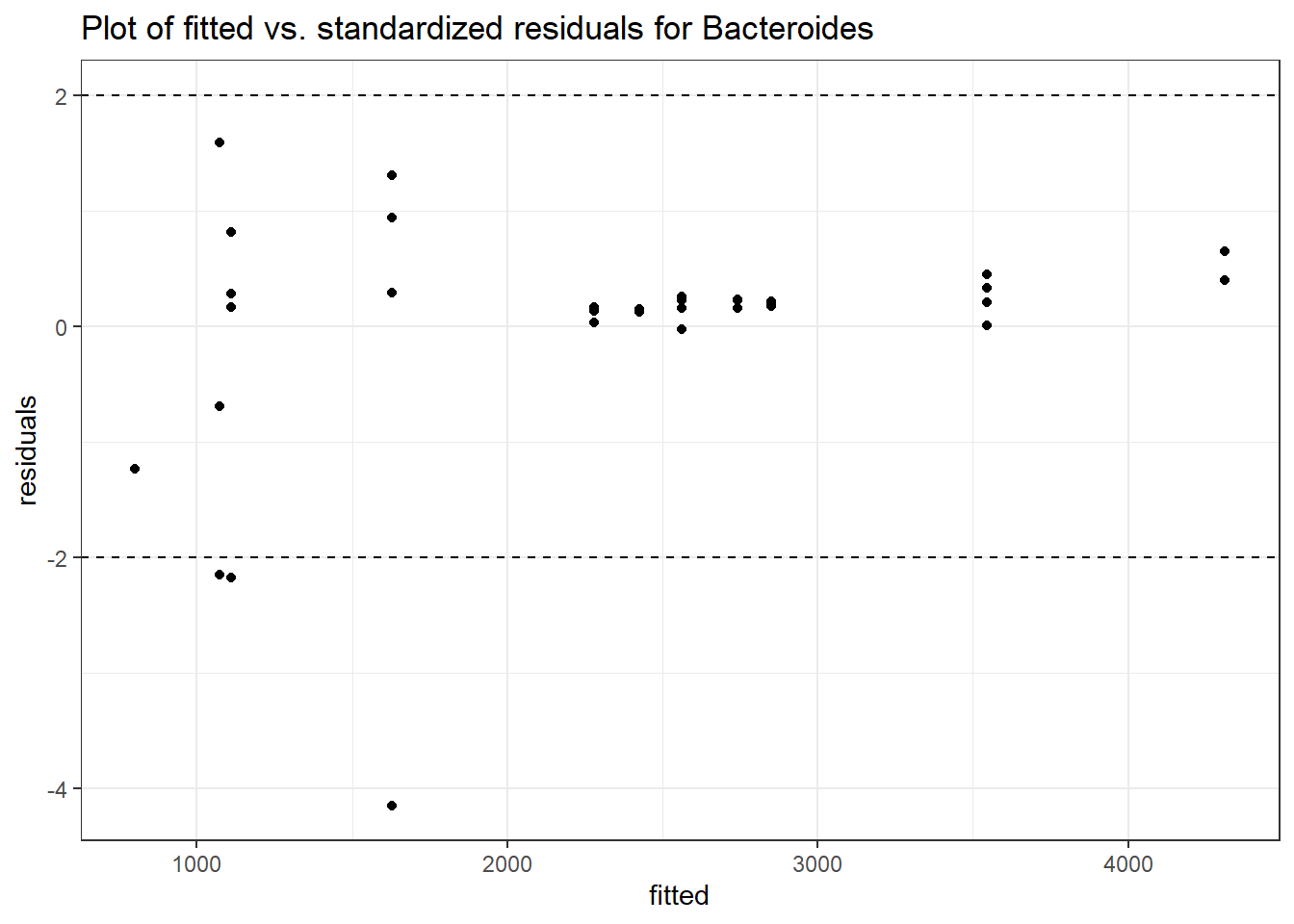
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.62374 0.1729 44.09211 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.53946
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00534952 (tol = 0.002, component 1)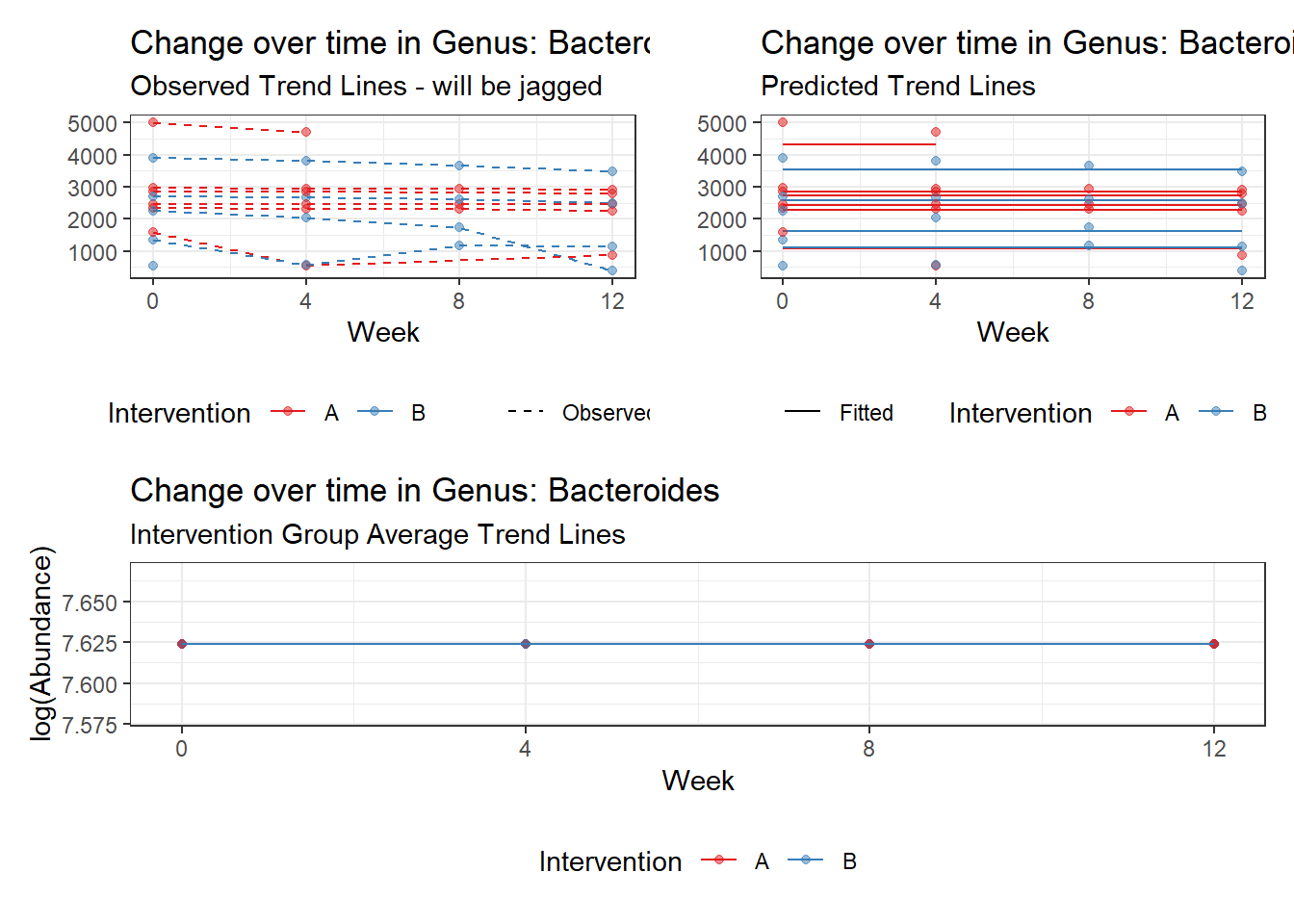
Intraclass Correlation (ICC): 0.3772285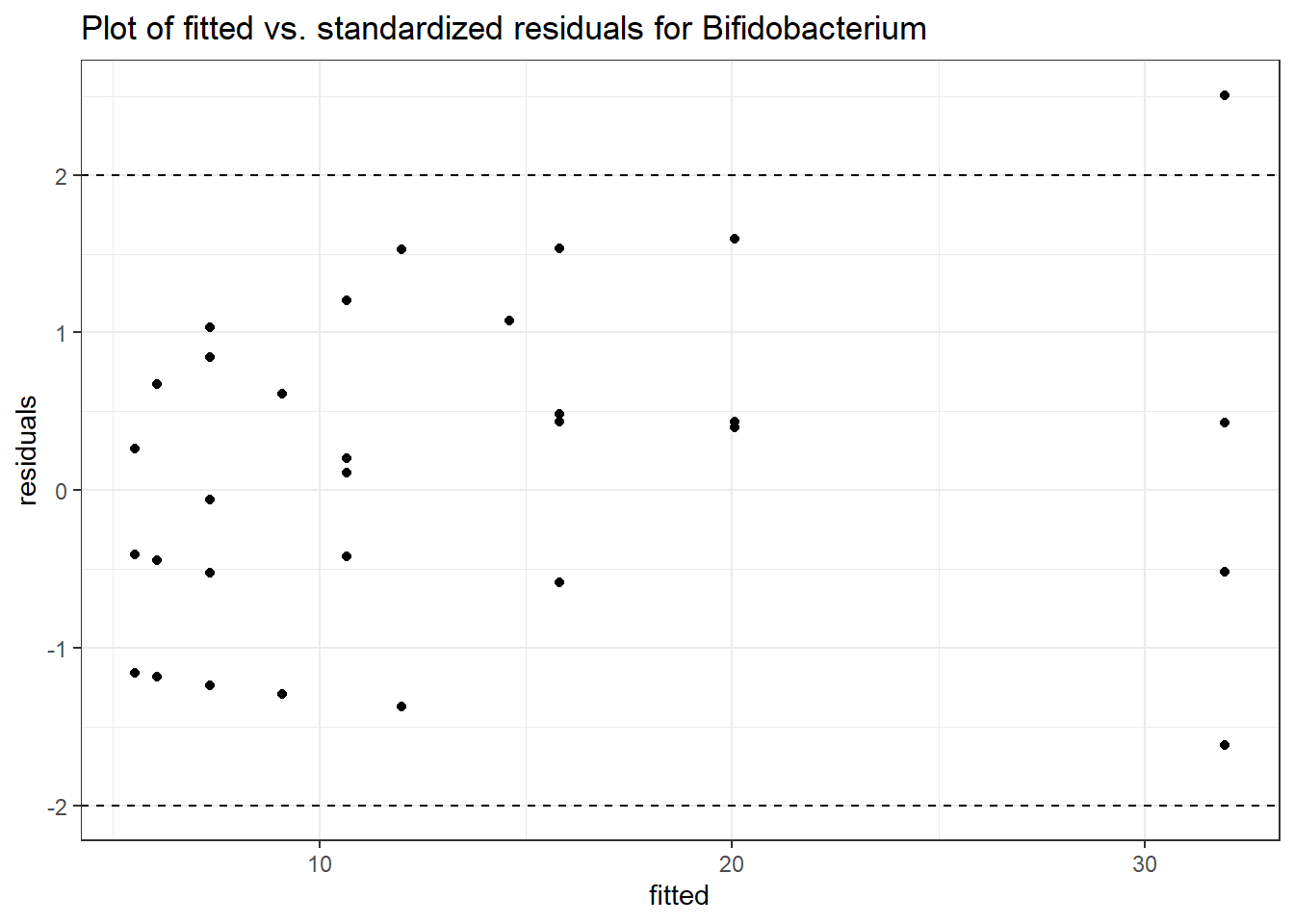
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.40237 0.39559 6.07286 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.77828
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinom
Intraclass Correlation (ICC): 0.4265957
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 1.08476 0.53115 2.04228 0.04112
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.86254
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinom
Intraclass Correlation (ICC): 0.04921848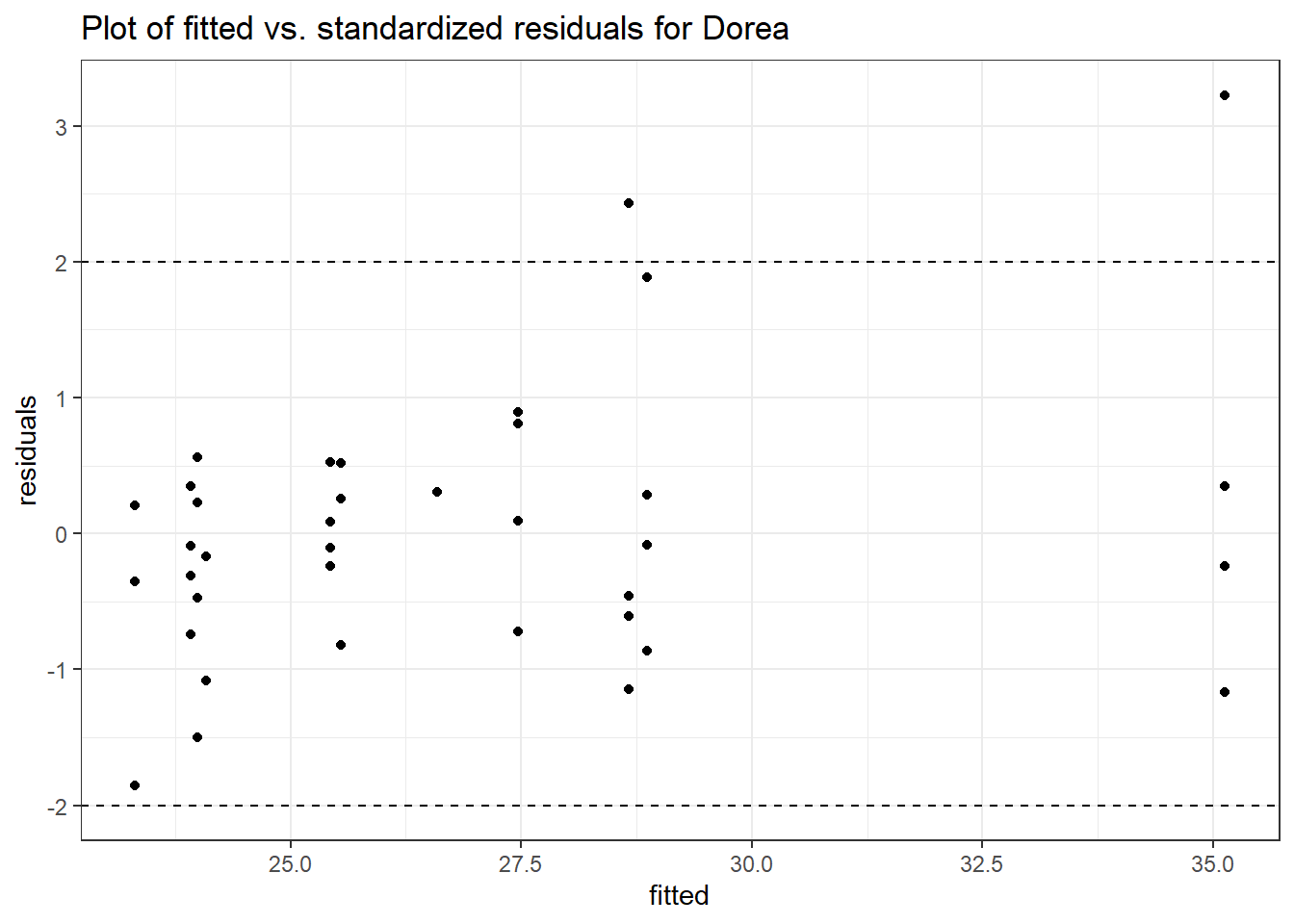
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.2751 0.14661 22.33891 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.22752
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinomboundary (singular) fit: see ?isSingular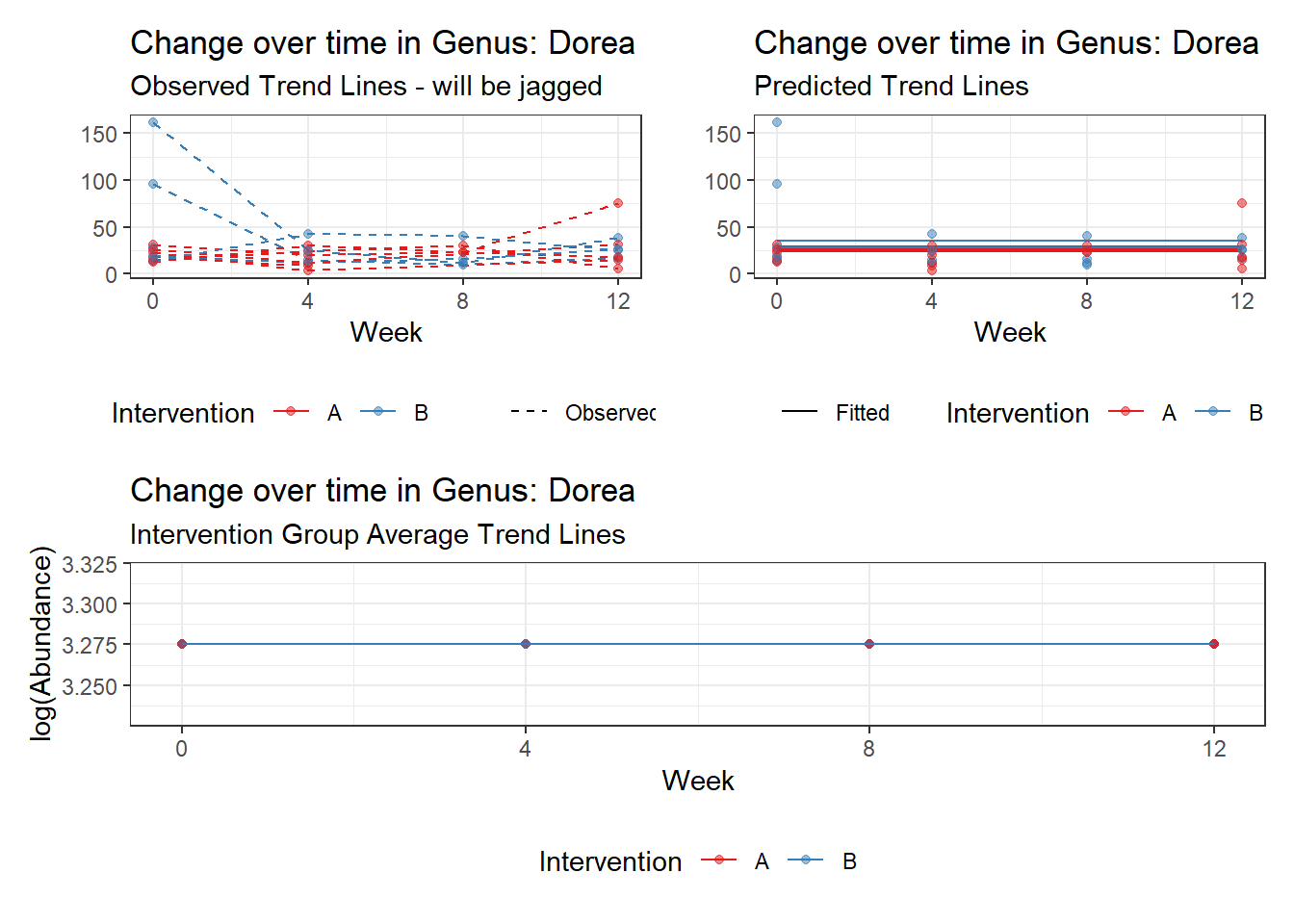
Intraclass Correlation (ICC): 0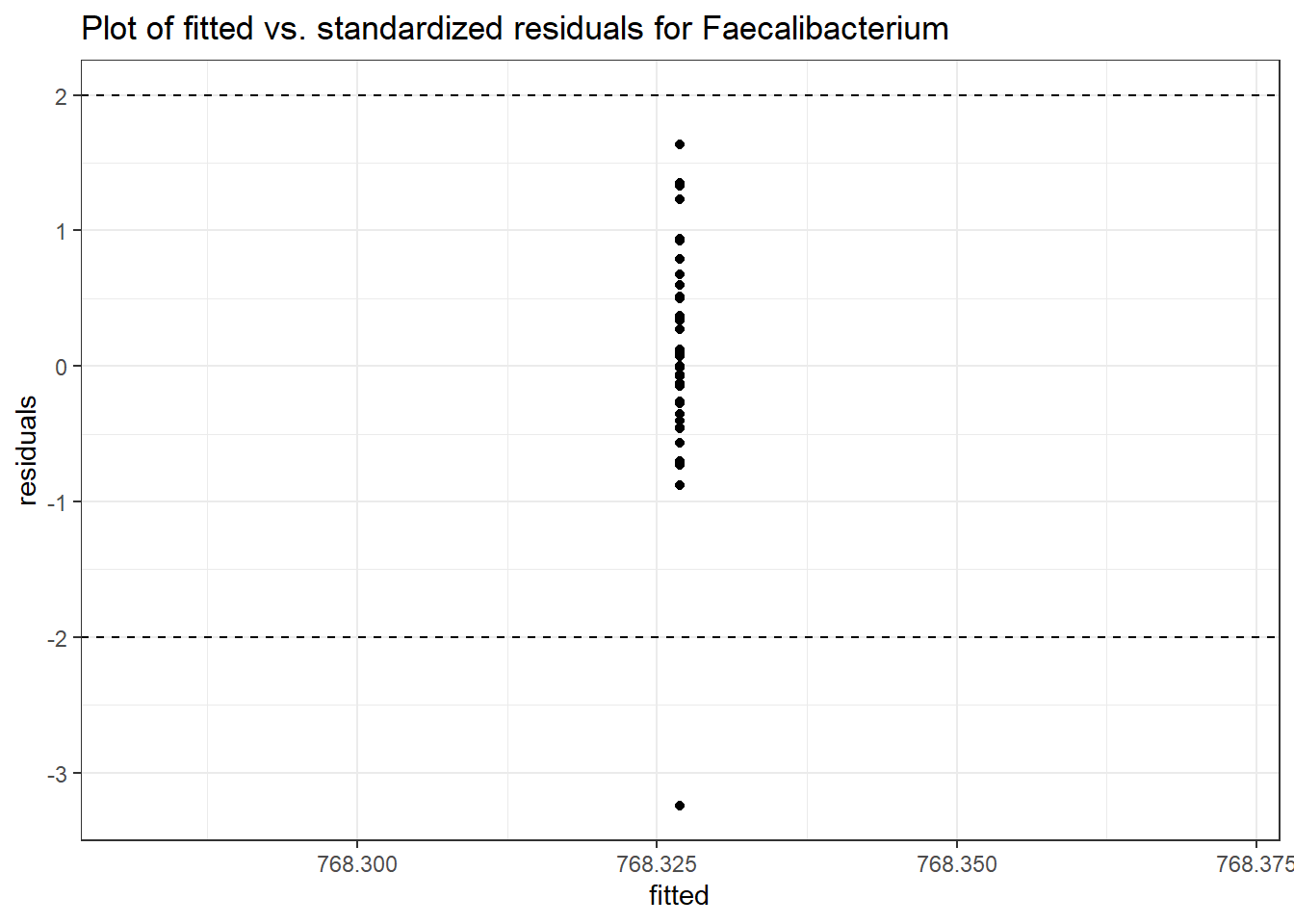
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.64422 0.15883 41.83259 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reached
Warning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, : NaNs
produced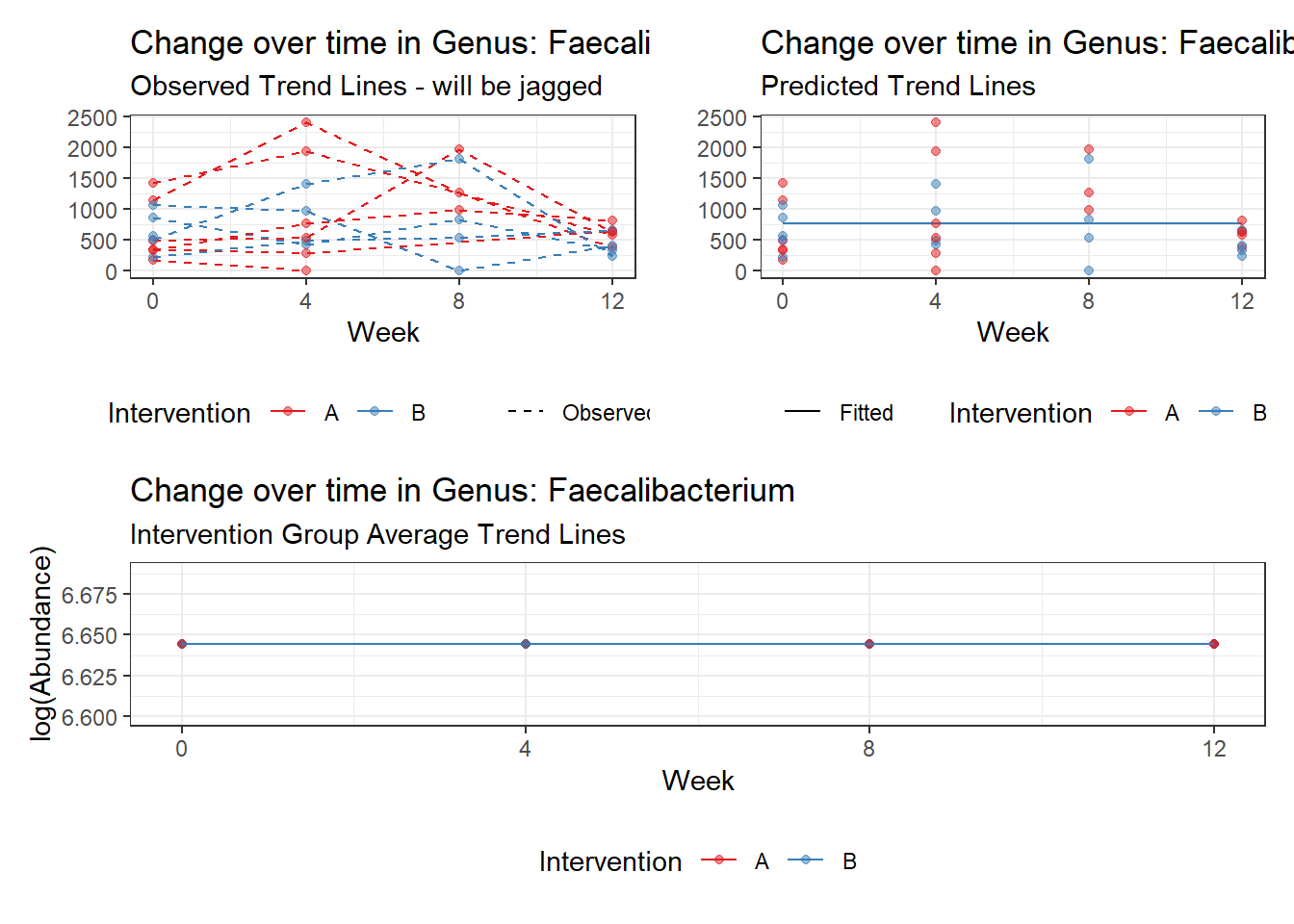
Intraclass Correlation (ICC): 0.3651309
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 3.9552 0.23075 17.14058 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.75837
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reached
Warning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, : NaNs
produced
Intraclass Correlation (ICC): 0.8657244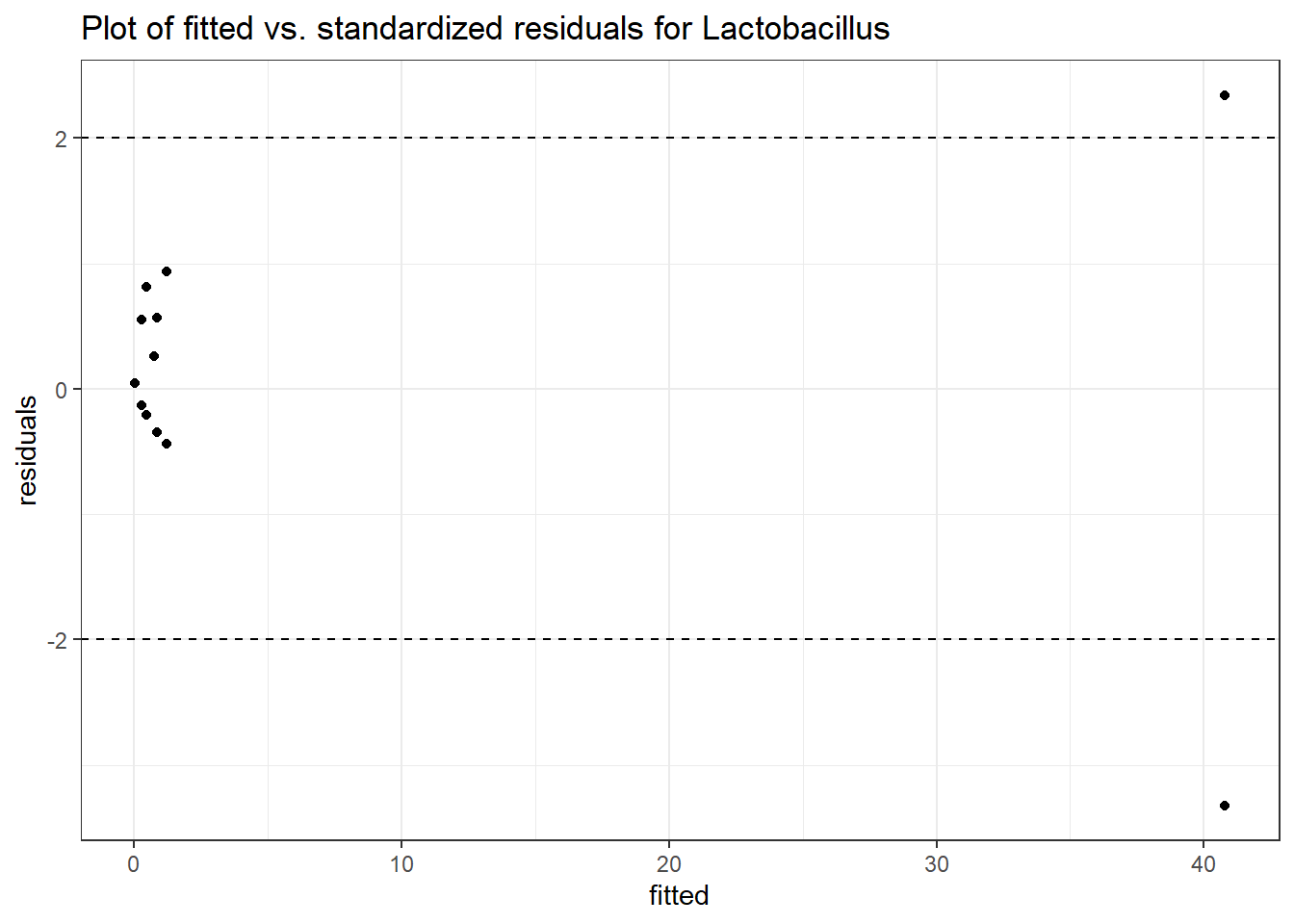
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus -1.70194 1.01557 -1.67585 0.09377
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.5392
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinomboundary (singular) fit: see ?isSingular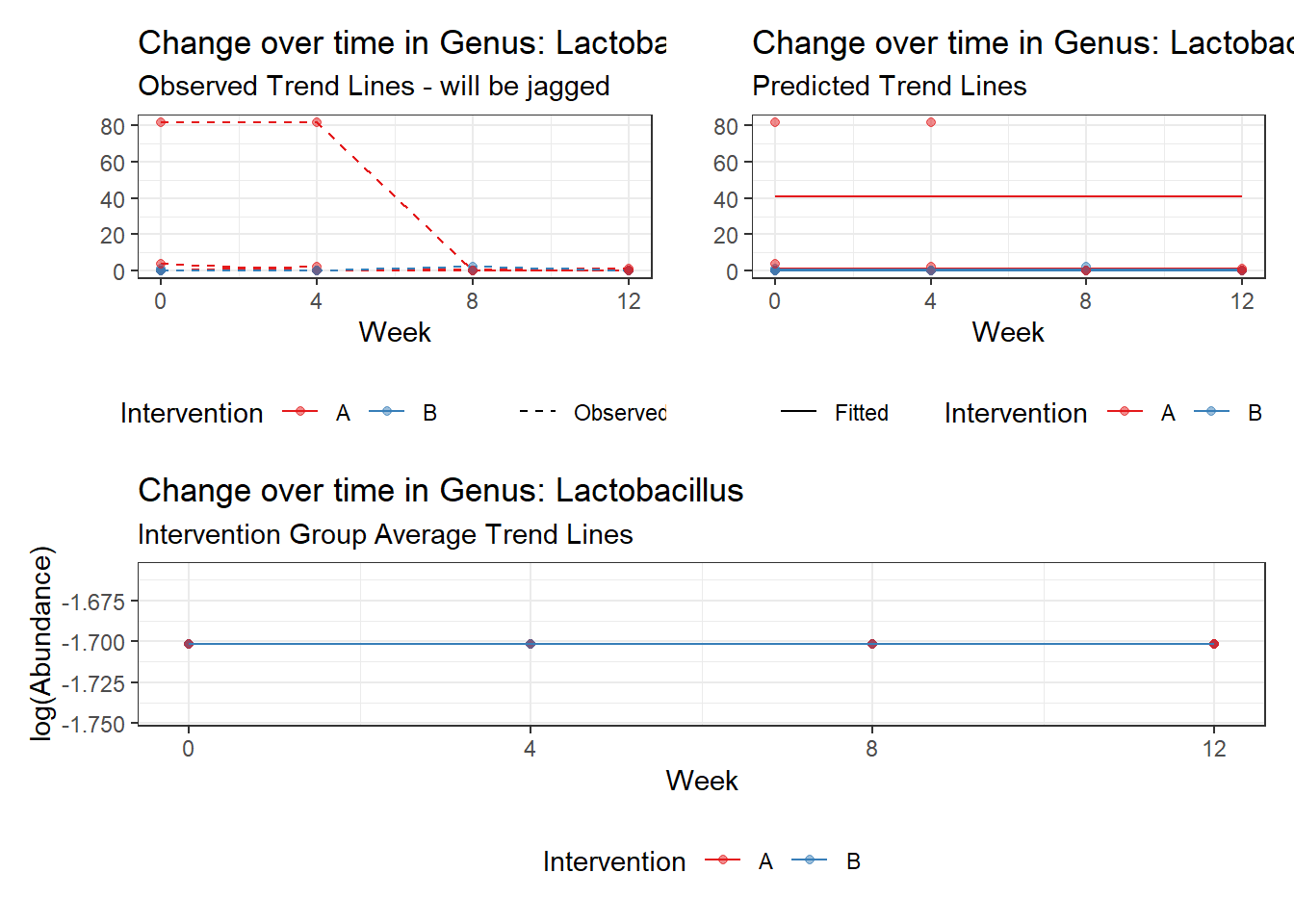
Intraclass Correlation (ICC): 0
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella 0.05264 0.53632 0.09815 0.92181
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0141497 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 0
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 3.14369 0.24881 12.63477 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + (1 | SubjectID)
<environment: 0x0000000038f39bc8>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0087568 (tol = 0.002, component 1)Error in factory(refitNB, types = c("message", "warning"))(lastfit, theta = exp(t), : pwrssUpdate did not converge in (maxit) iterations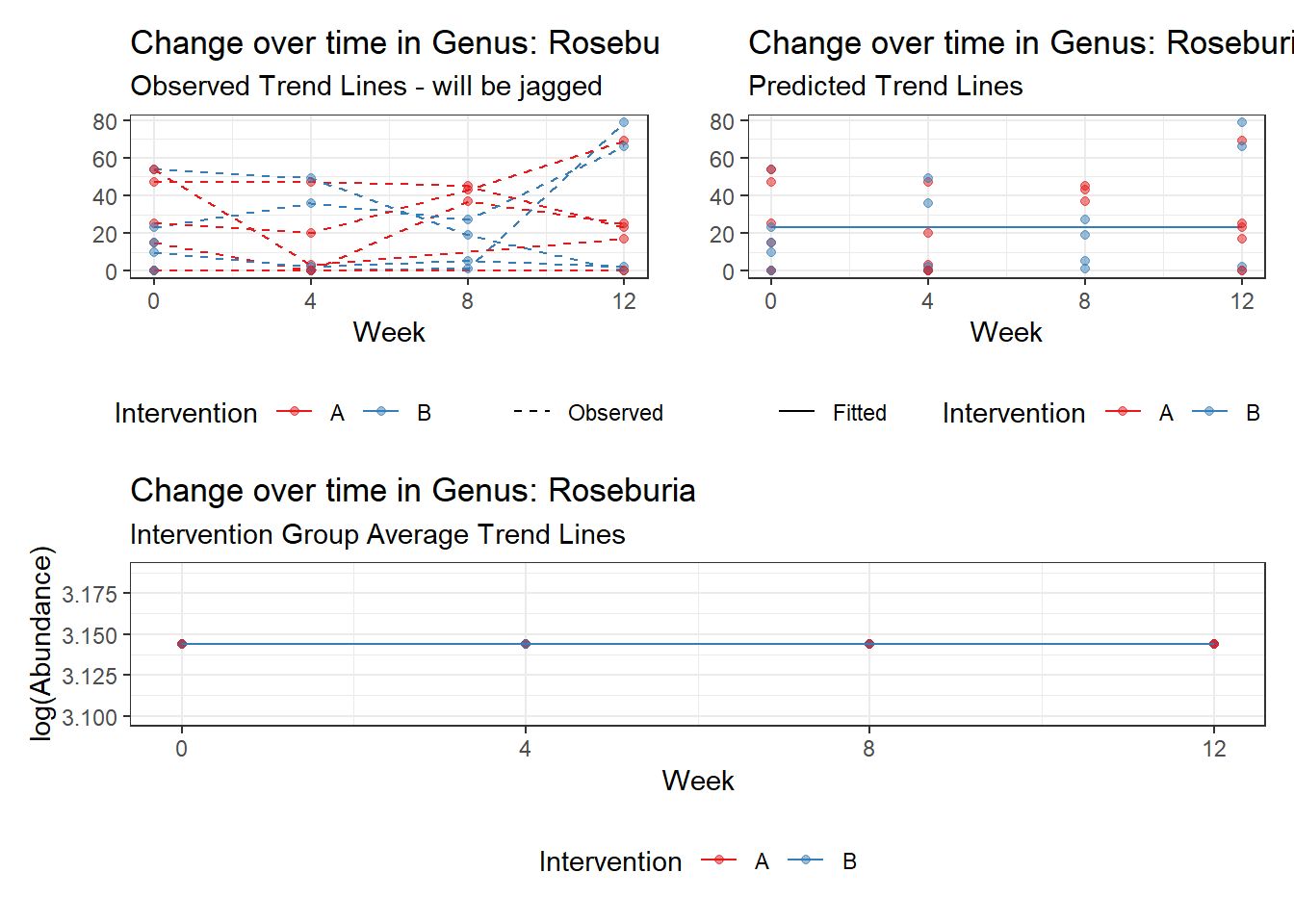
Fixed Effect of Time
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
genus.fit1 <- glmm_microbiome(mydata=microbiome_data,model.number=1,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + time + (1|SubjectID)")
#########################################
#########################################
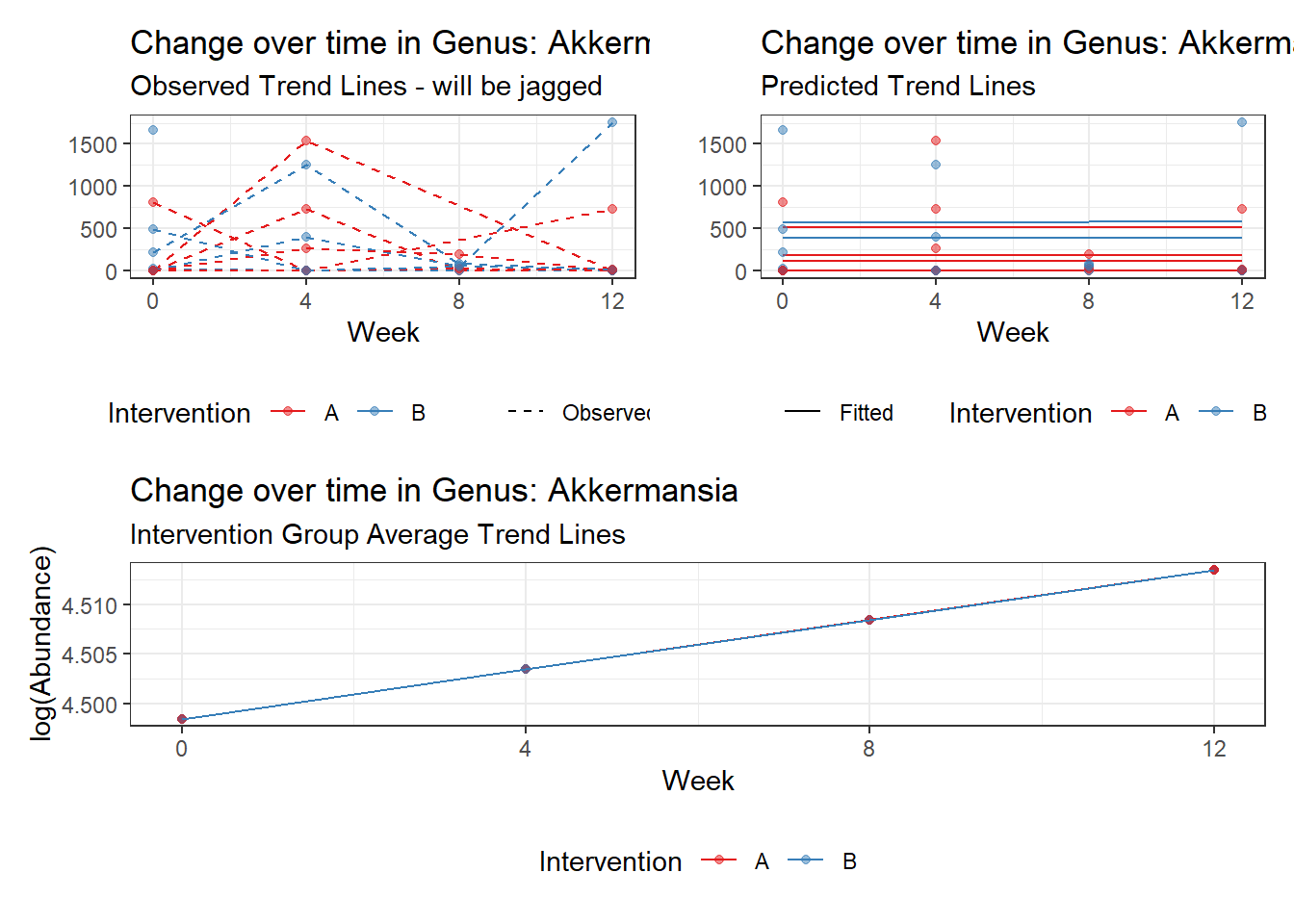
Model: Akkermansia ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.8607546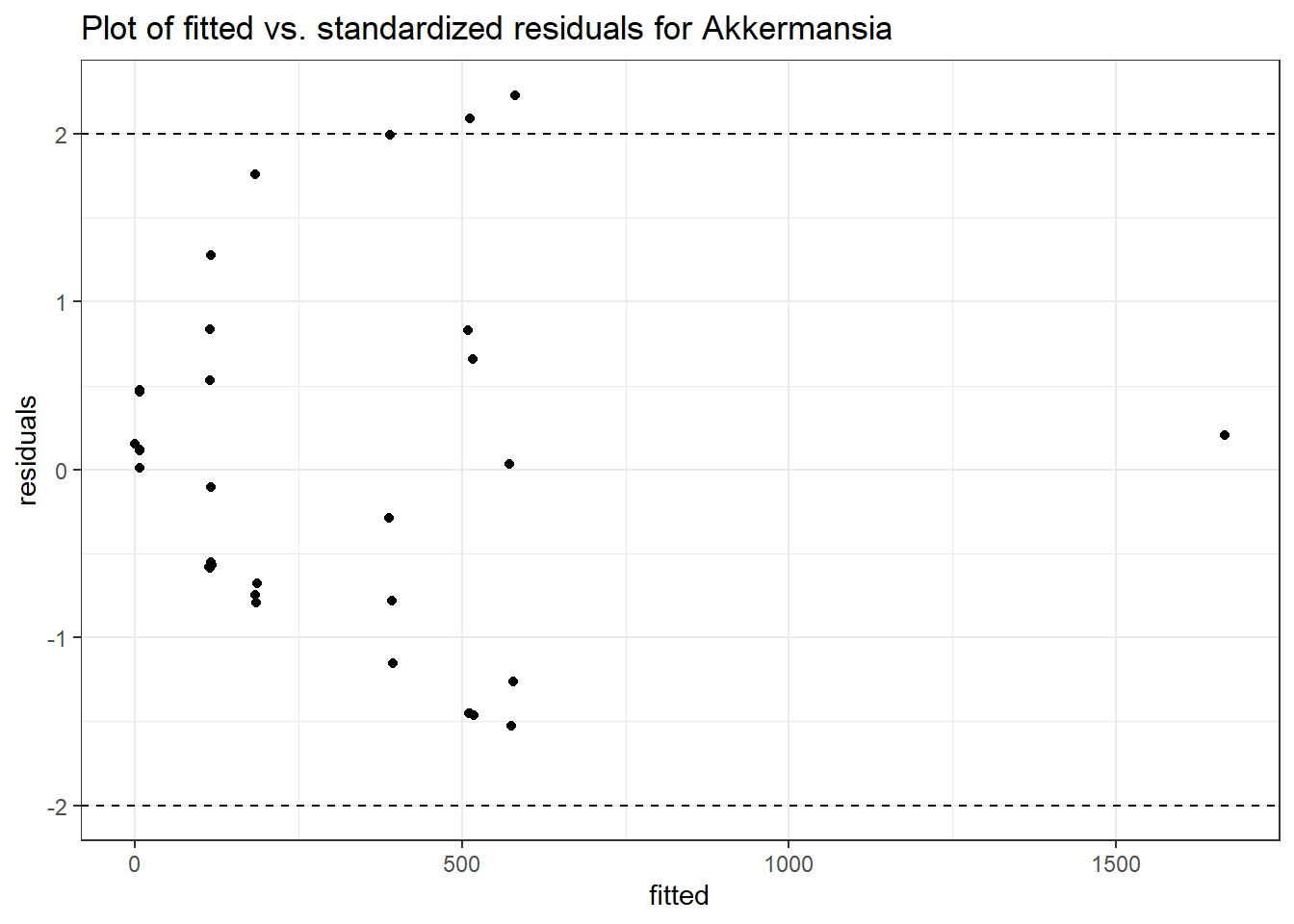
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 4.49846 0.75628 5.94816 0.00000
time Akkermansia 0.00500 0.00920 0.54317 0.58701
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4863
Significance test not available for random effects
#########################################
#########################################
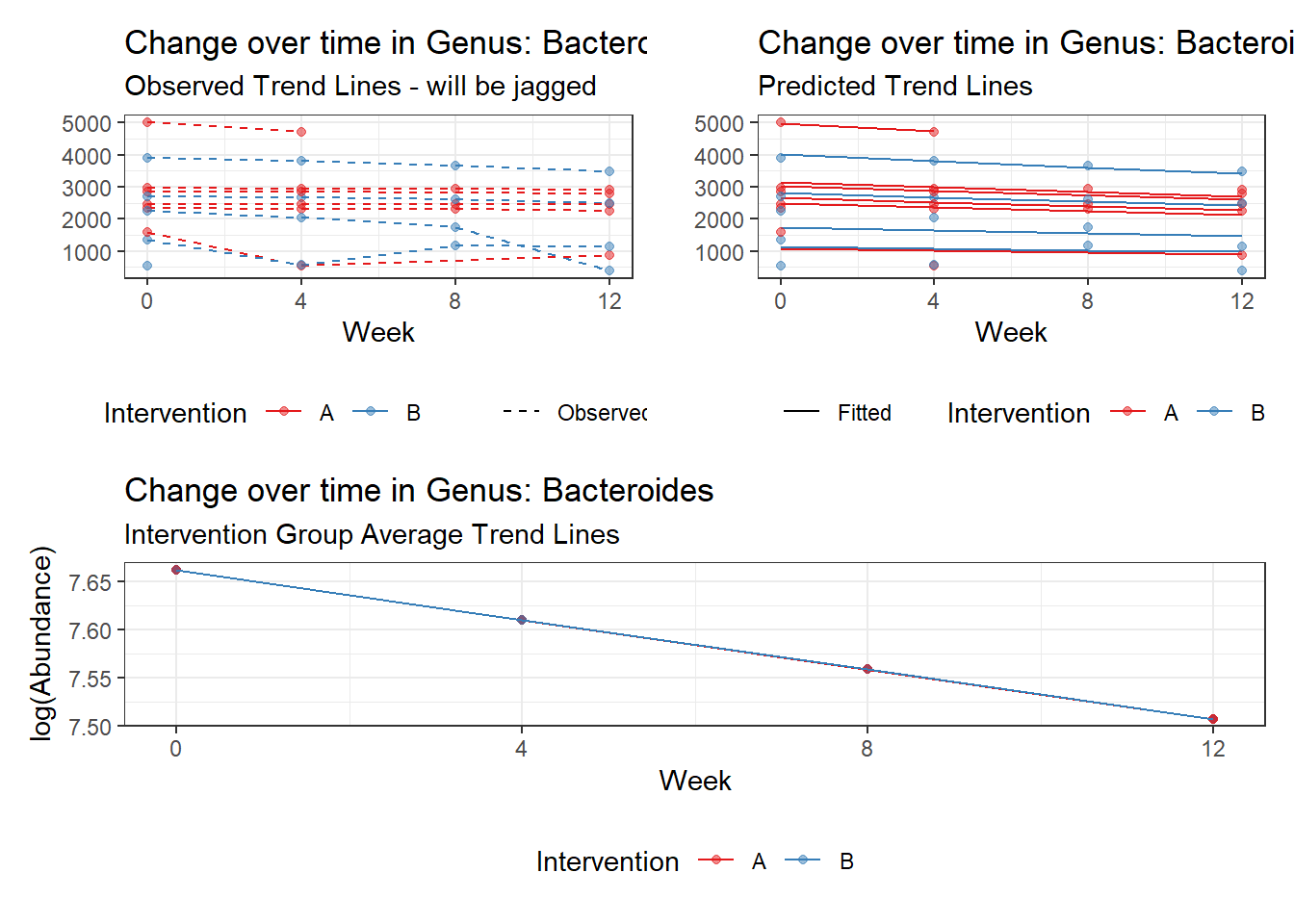
Model: Bacteroides ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.2823366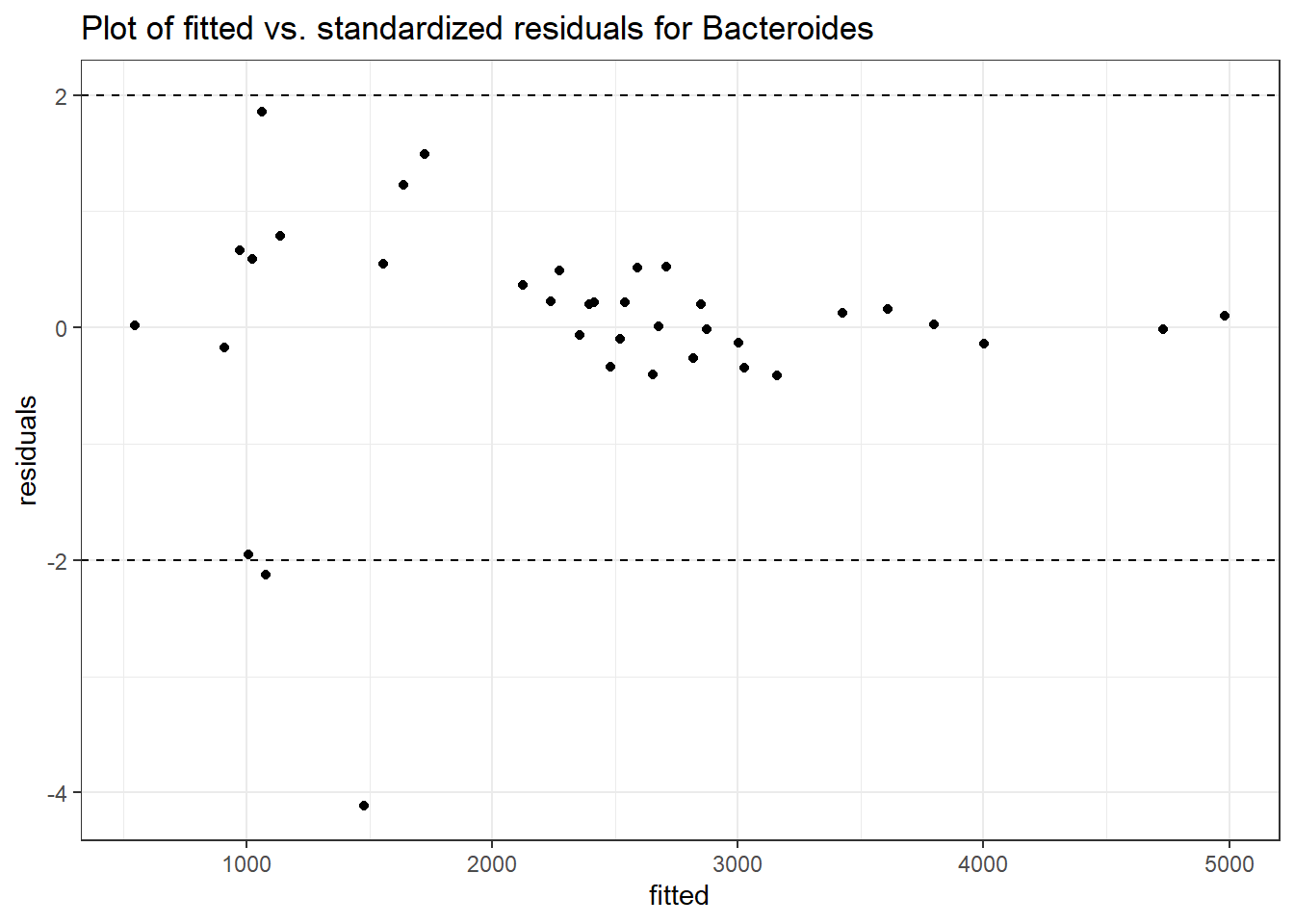
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.66234 0.18922 40.49392 0
time Bacteroides -0.05161 0.00312 -16.55748 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.62722
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.5684684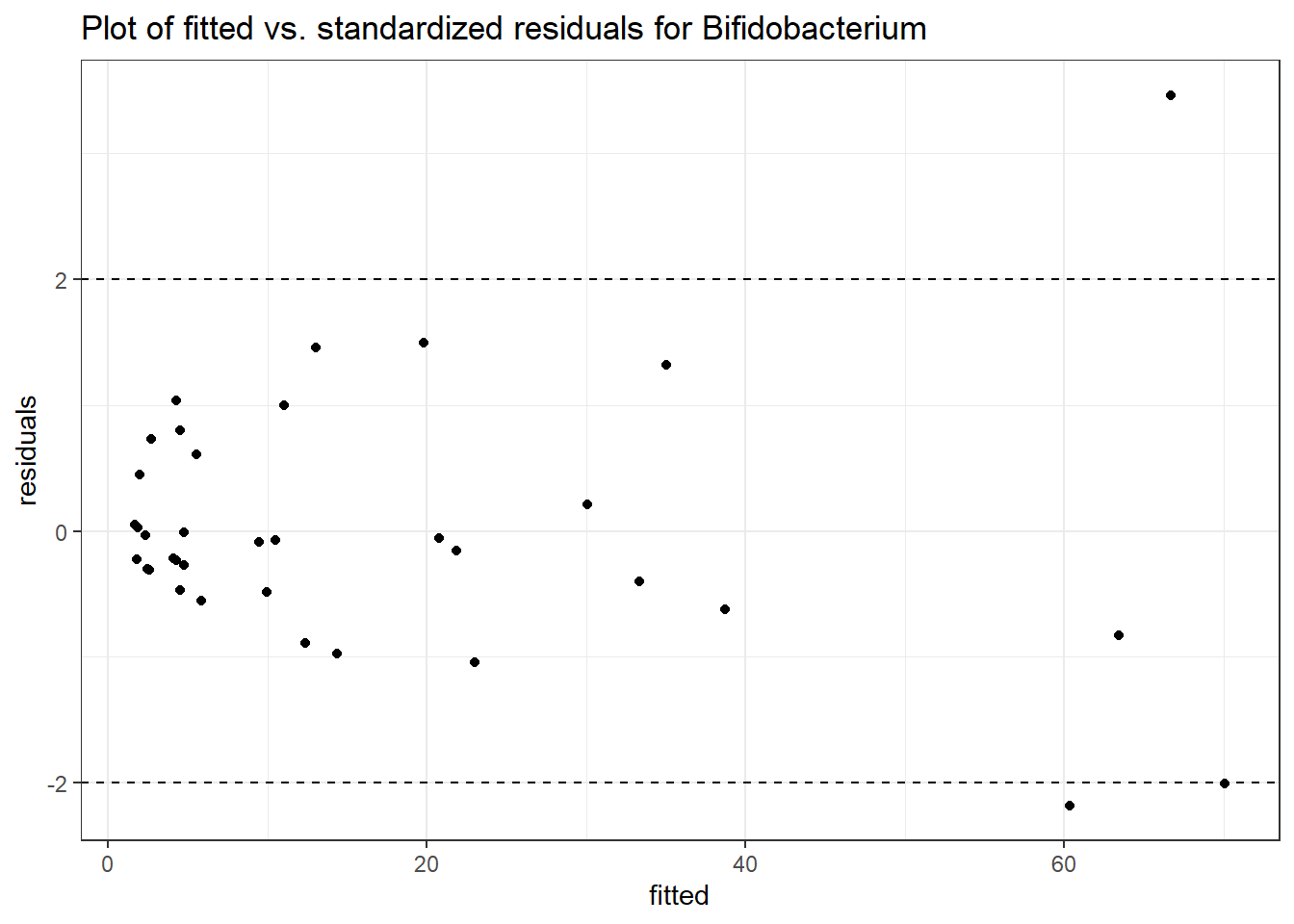

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.21950 0.35564 6.24077 0.00000
time Bifidobacterium 0.04988 0.03554 1.40363 0.16043
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.1477
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.6511505
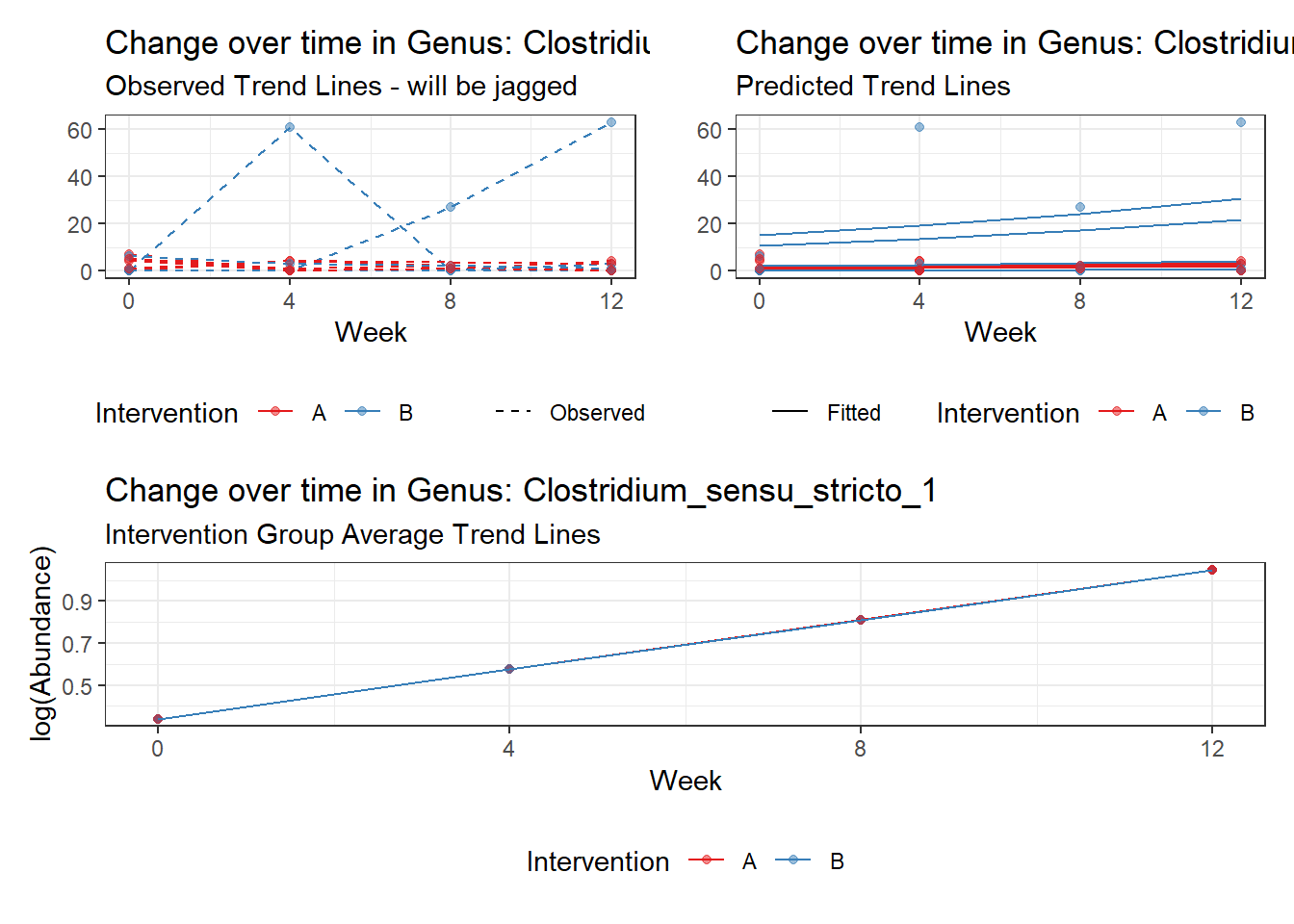
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.34008 0.46124 0.73730 0.46094
time Clostridium_sensu_stricto_1 0.23592 0.06296 3.74708 0.00018
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.3662
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.1606926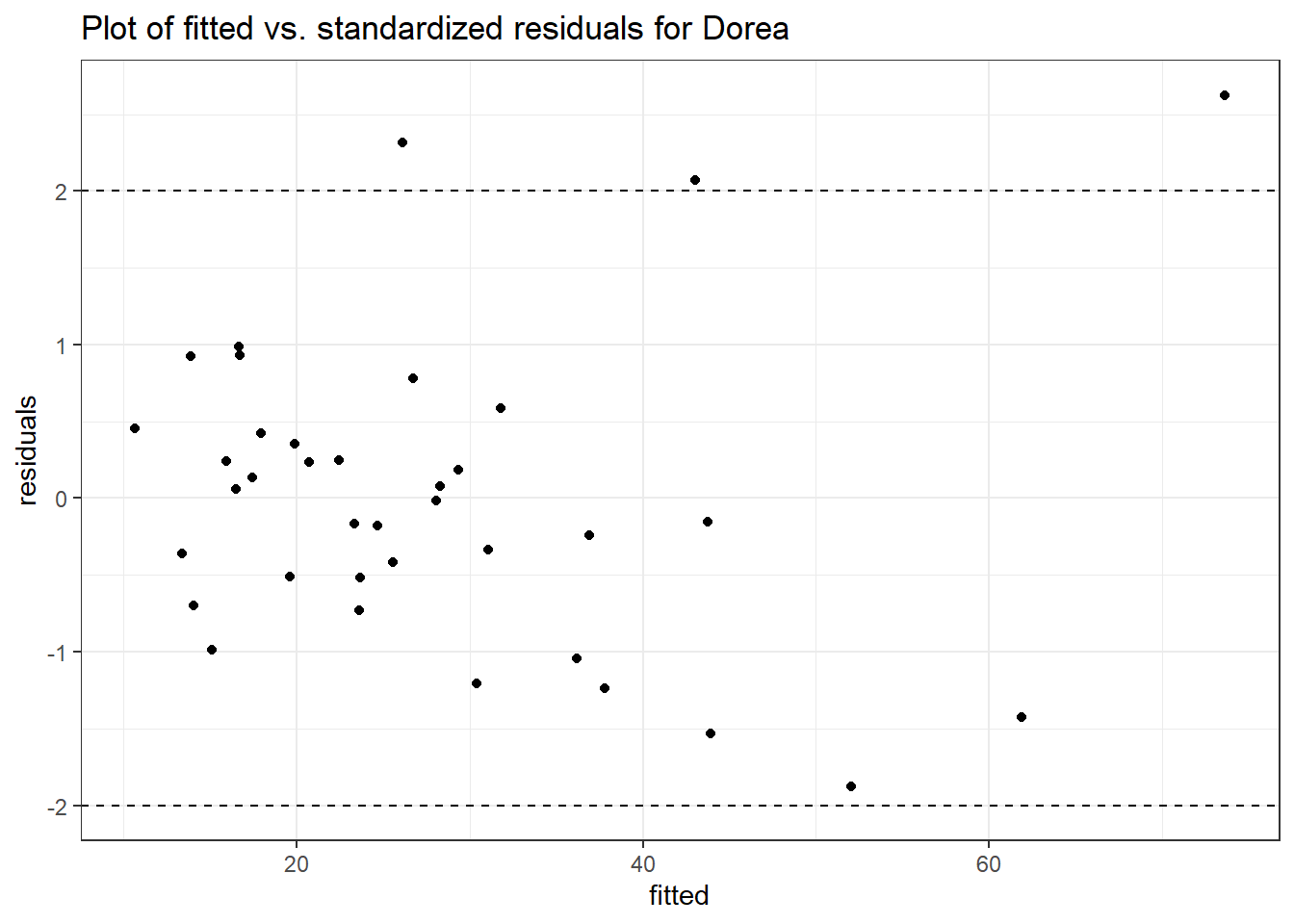
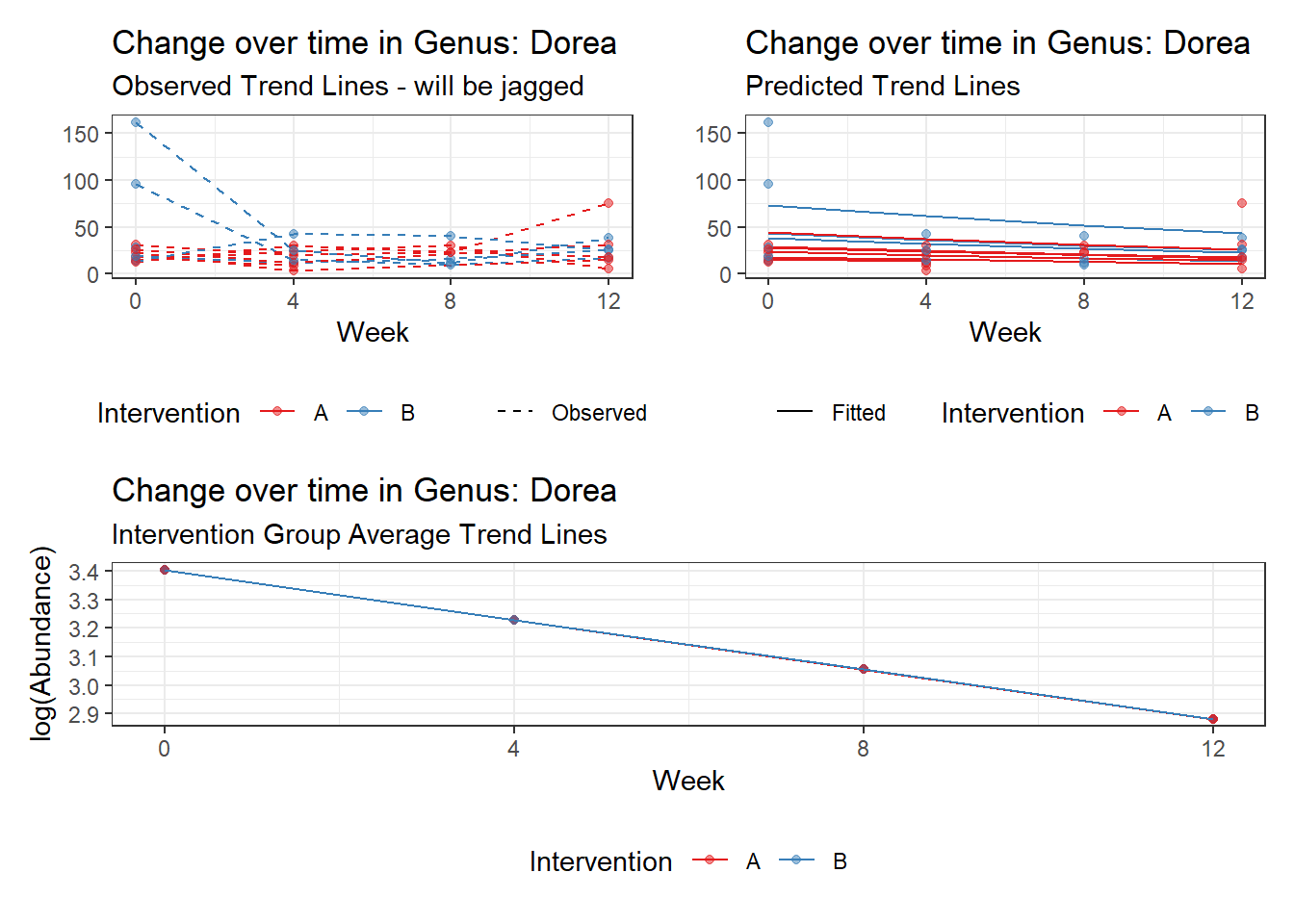
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.40303 0.14054 24.21326 0
time Dorea -0.17374 0.02858 -6.07987 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.43756
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.3447693

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.50046 0.21900 29.68227 0
time Faecalibacterium -0.07486 0.00529 -14.16068 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.72538
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.3575527

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 3.79918 0.22840 16.63358 0
time Lachnospira 0.11860 0.01758 6.74623 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.74602
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.8646958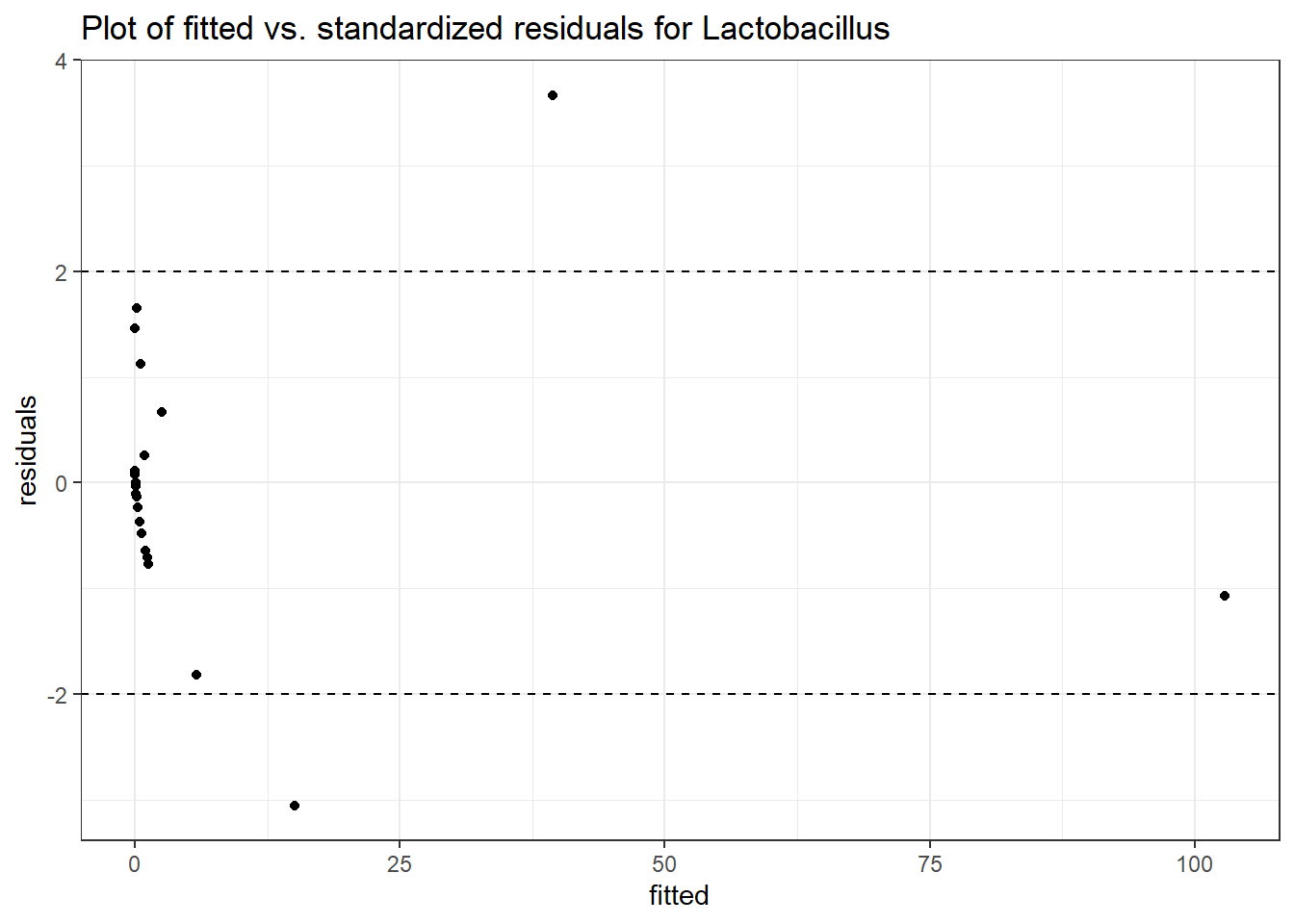
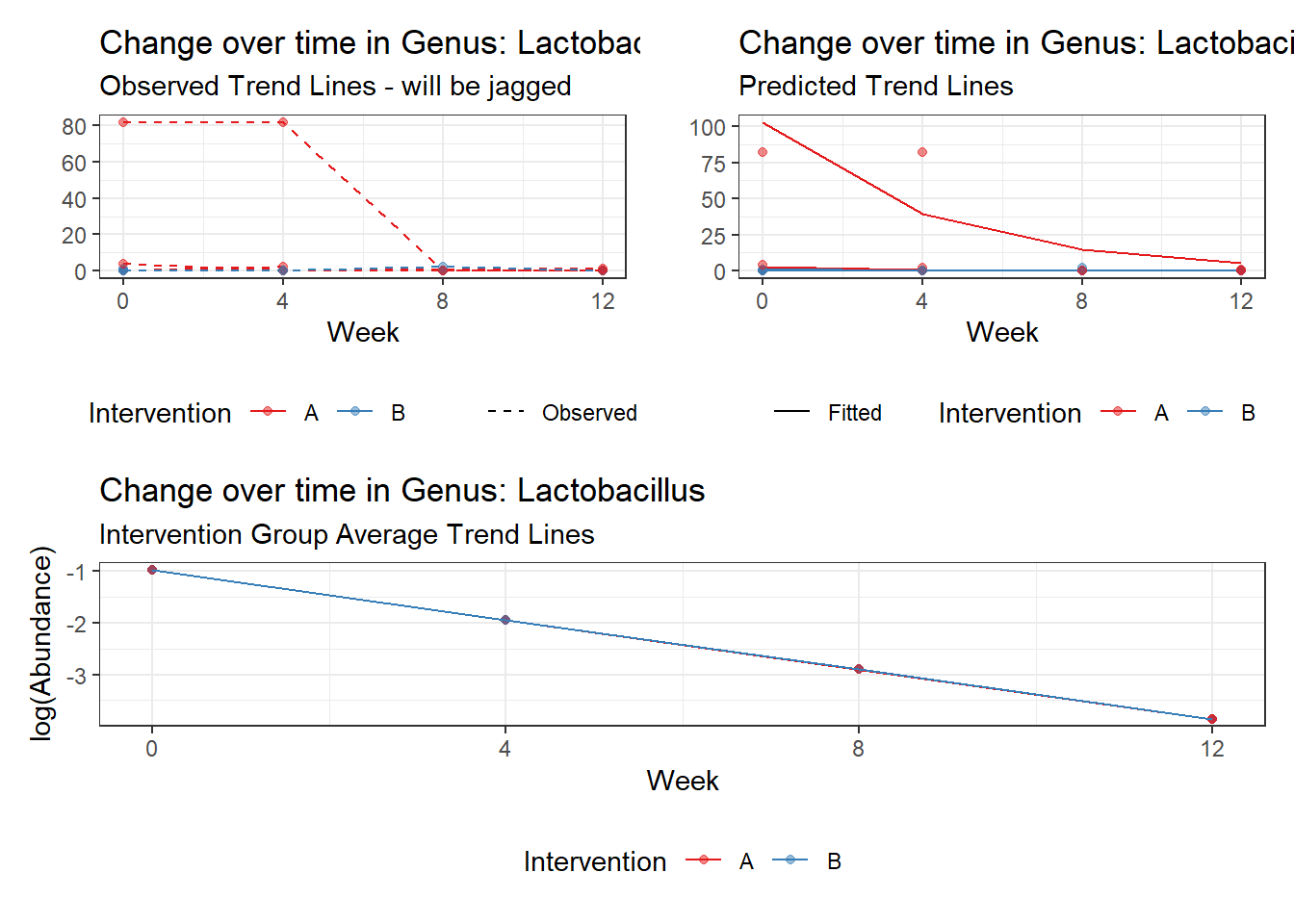
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus -0.97570 1.03272 -0.94479 0.34477
time Lactobacillus -0.96001 0.09524 -10.07970 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.528
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.7673368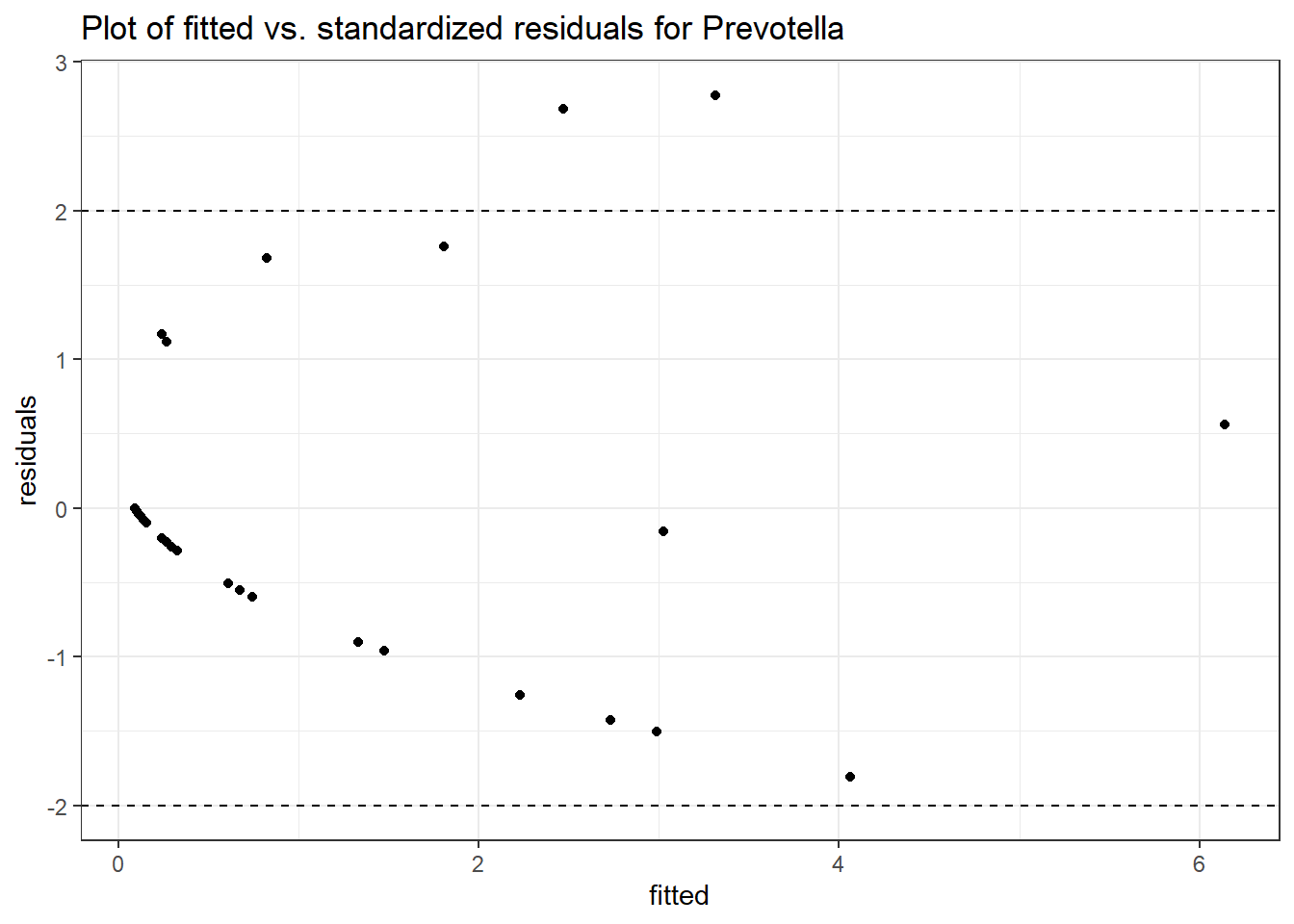

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.01146 0.73516 -1.37584 0.16887
time Prevotella 0.10231 0.14709 0.69556 0.48671
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.8161
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
Intraclass Correlation (ICC): 0.7558961
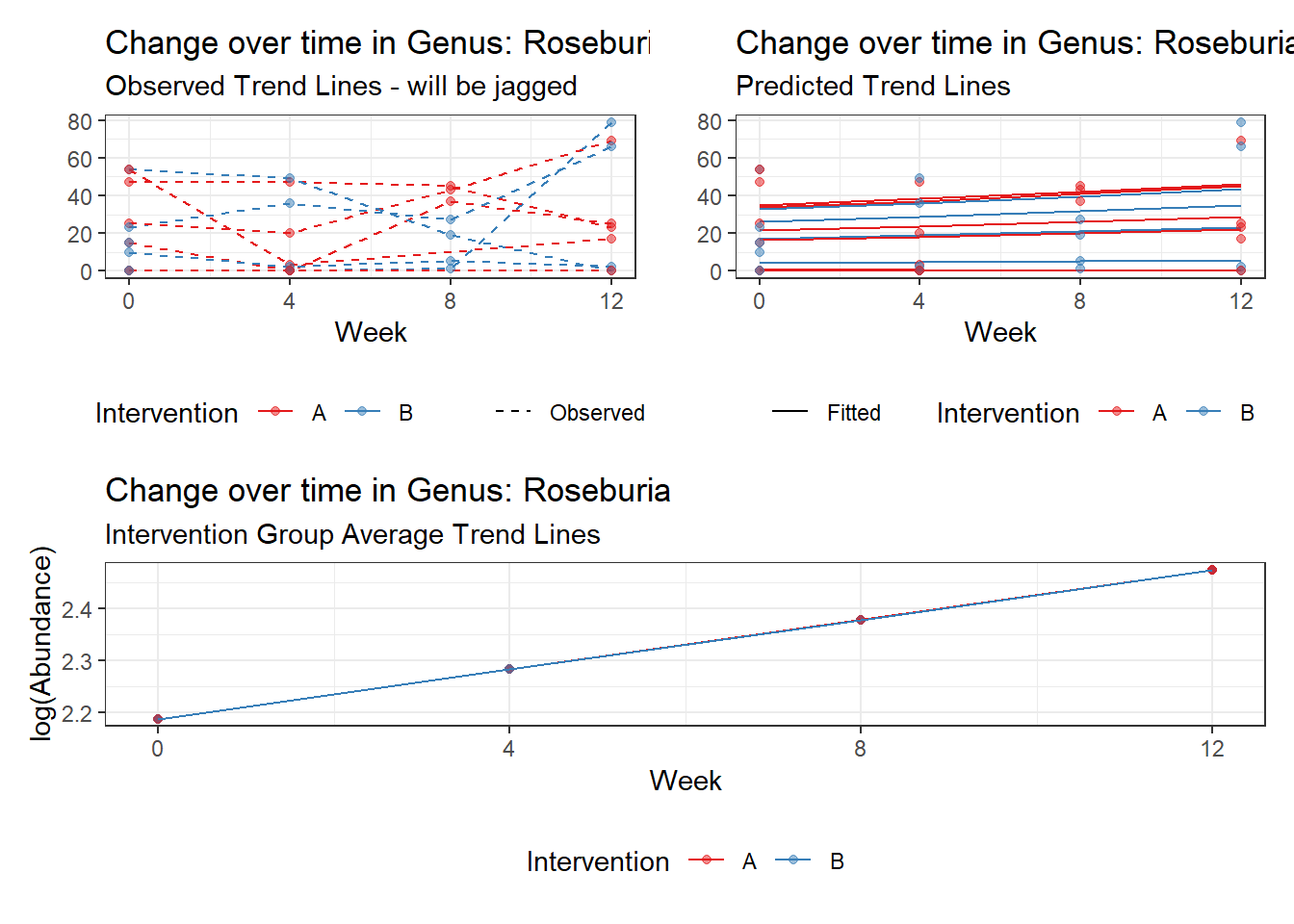
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 2.18772 0.55168 3.96554 0.00007
time Roseburia 0.09558 0.03045 3.13931 0.00169
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7597
Significance test not available for random effects
#########################################
#########################################
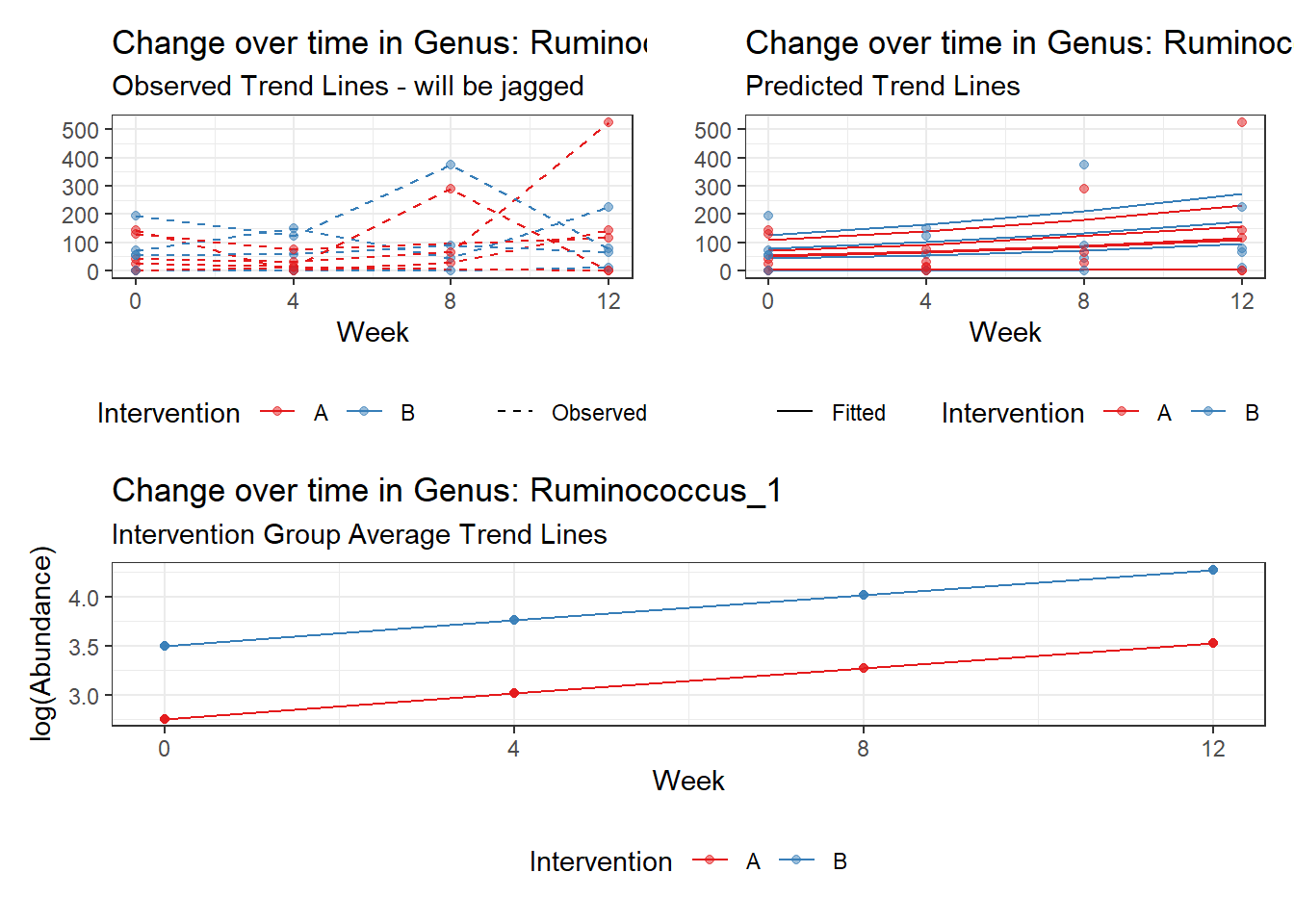
Model: Ruminococcus_1 ~ 1 + time + (1 | SubjectID)
<environment: 0x000000004bd68c10>
Link: poisson
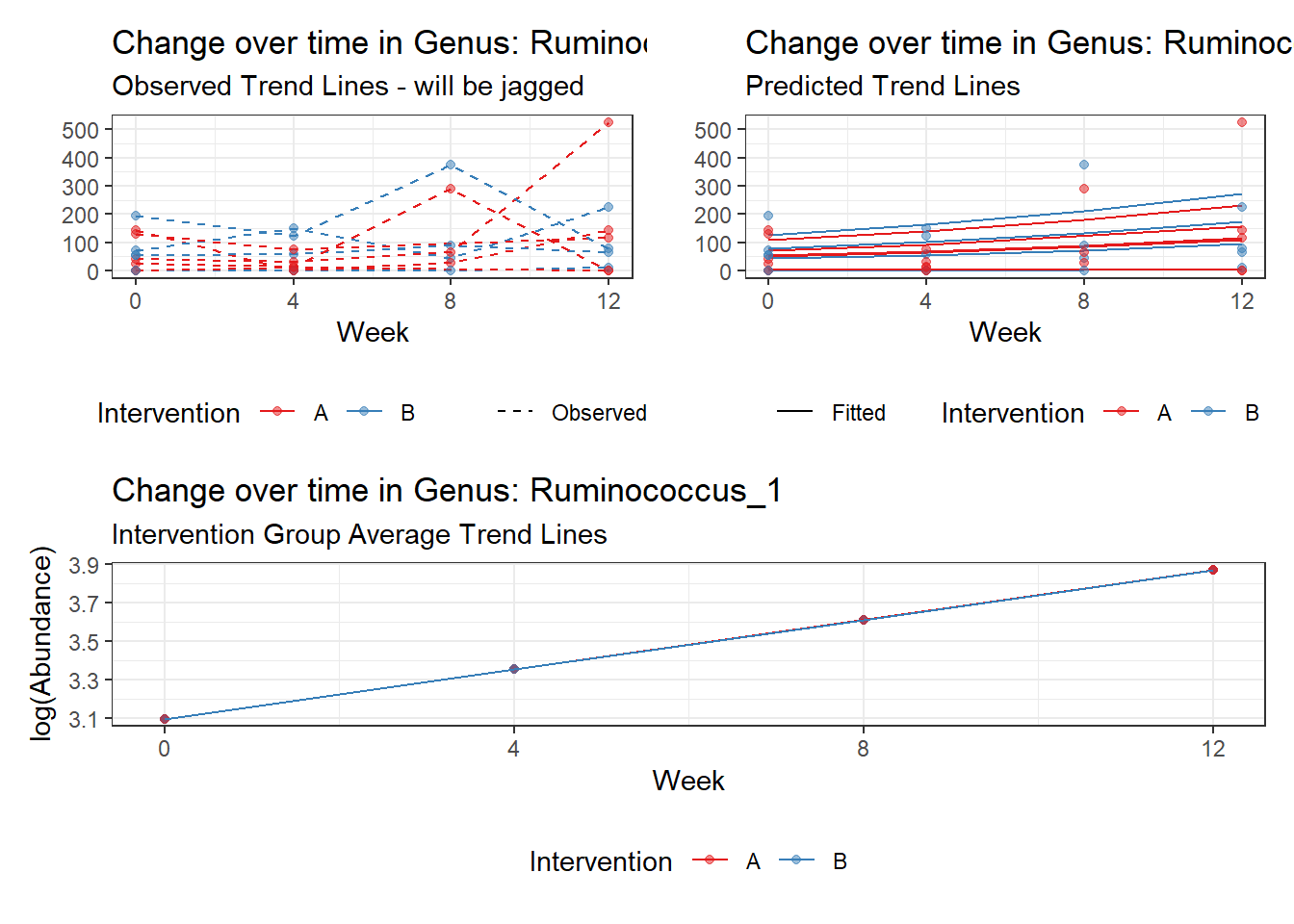
Intraclass Correlation (ICC): 0.7916593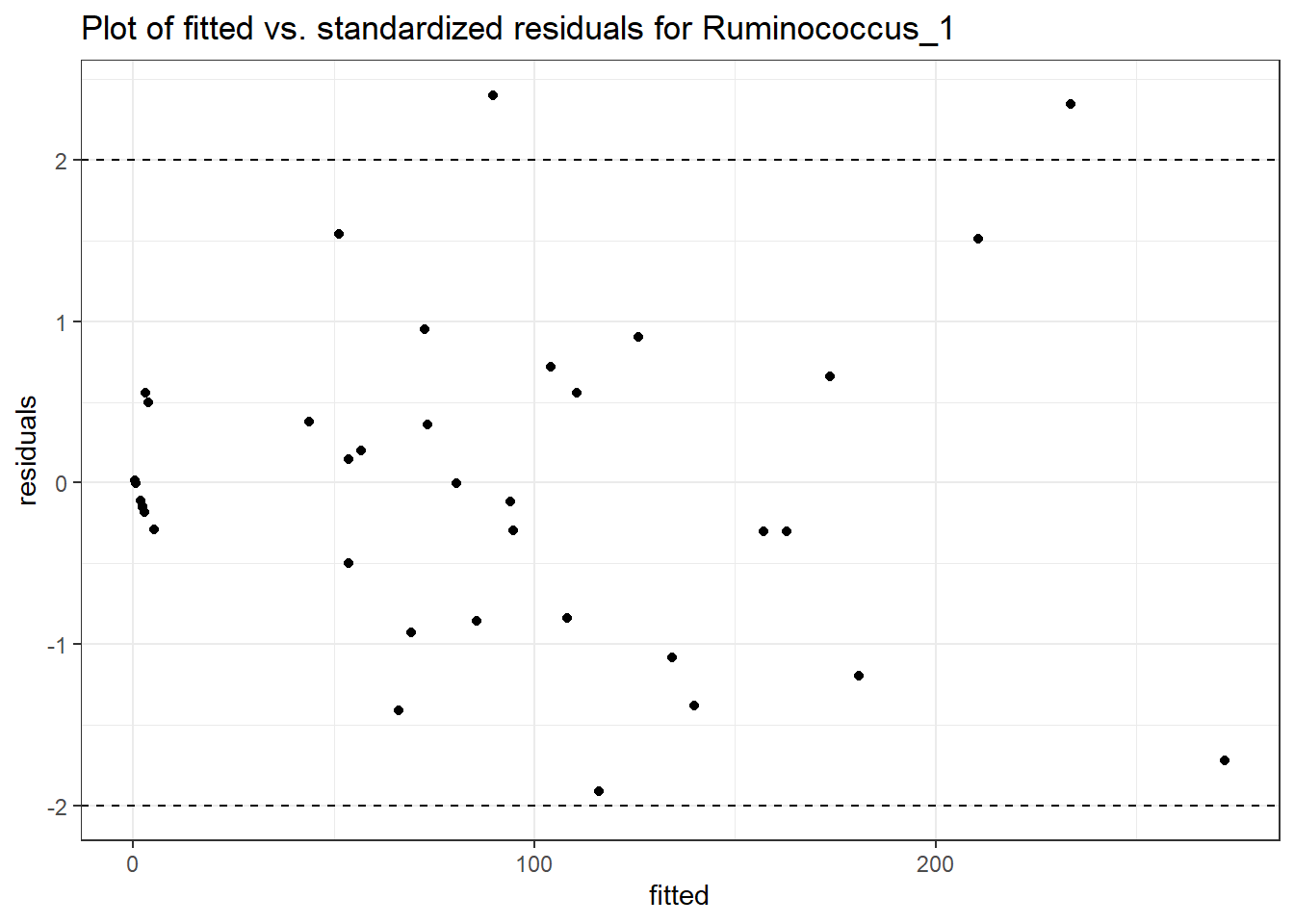
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 3.09853 0.59661 5.19355 0
time Ruminococcus_1 0.25673 0.01598 16.06222 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.9493
Significance test not available for random effectsgenus.fit1b <- glmm_microbiome(mydata=microbiome_data,model.number=1,
taxa.level="Genus", link="negbinom",
model="1 + time + (1|SubjectID)")
#########################################
#########################################
Model: Clostridium_innocuum_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reached
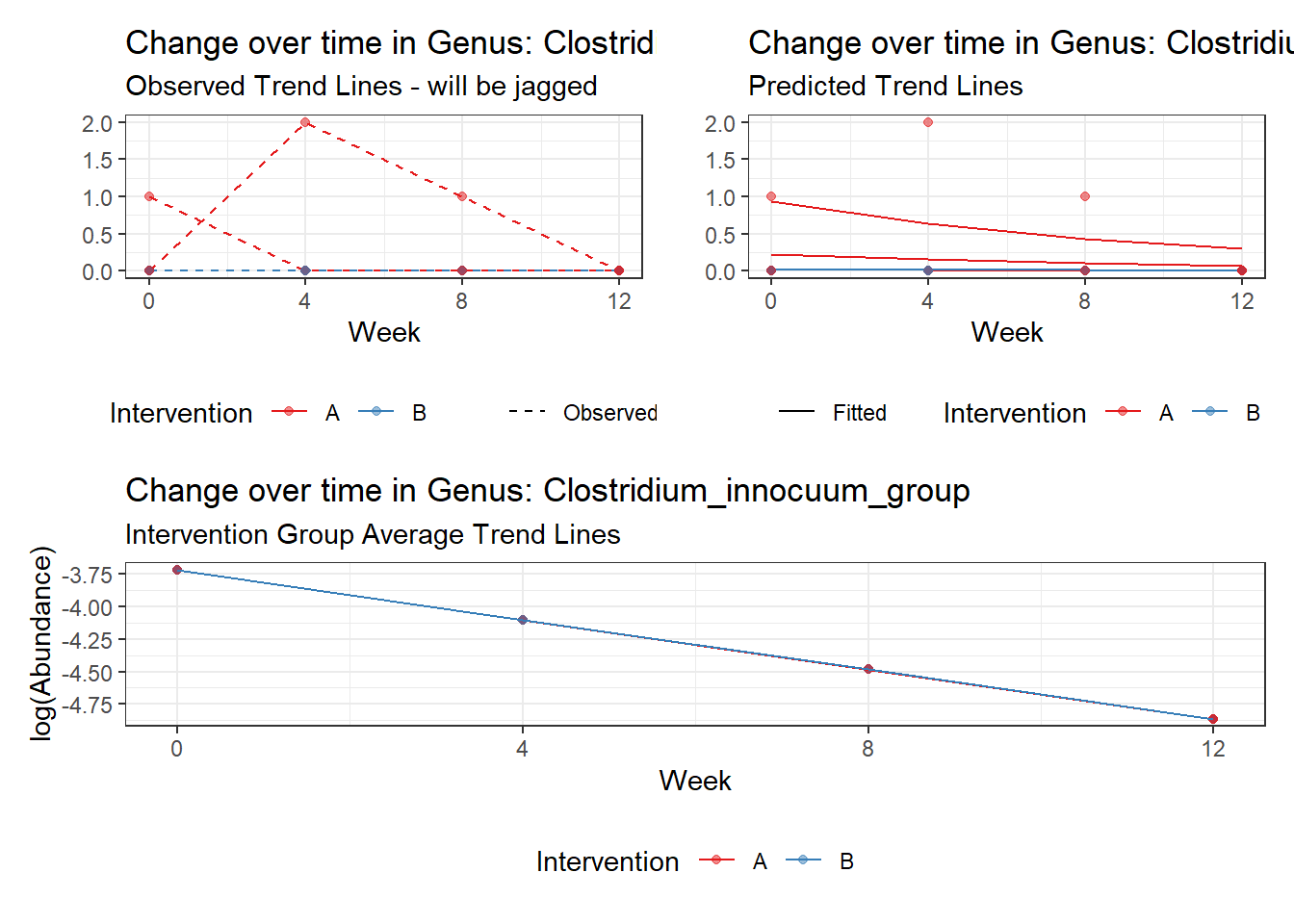
Intraclass Correlation (ICC): 0.8373419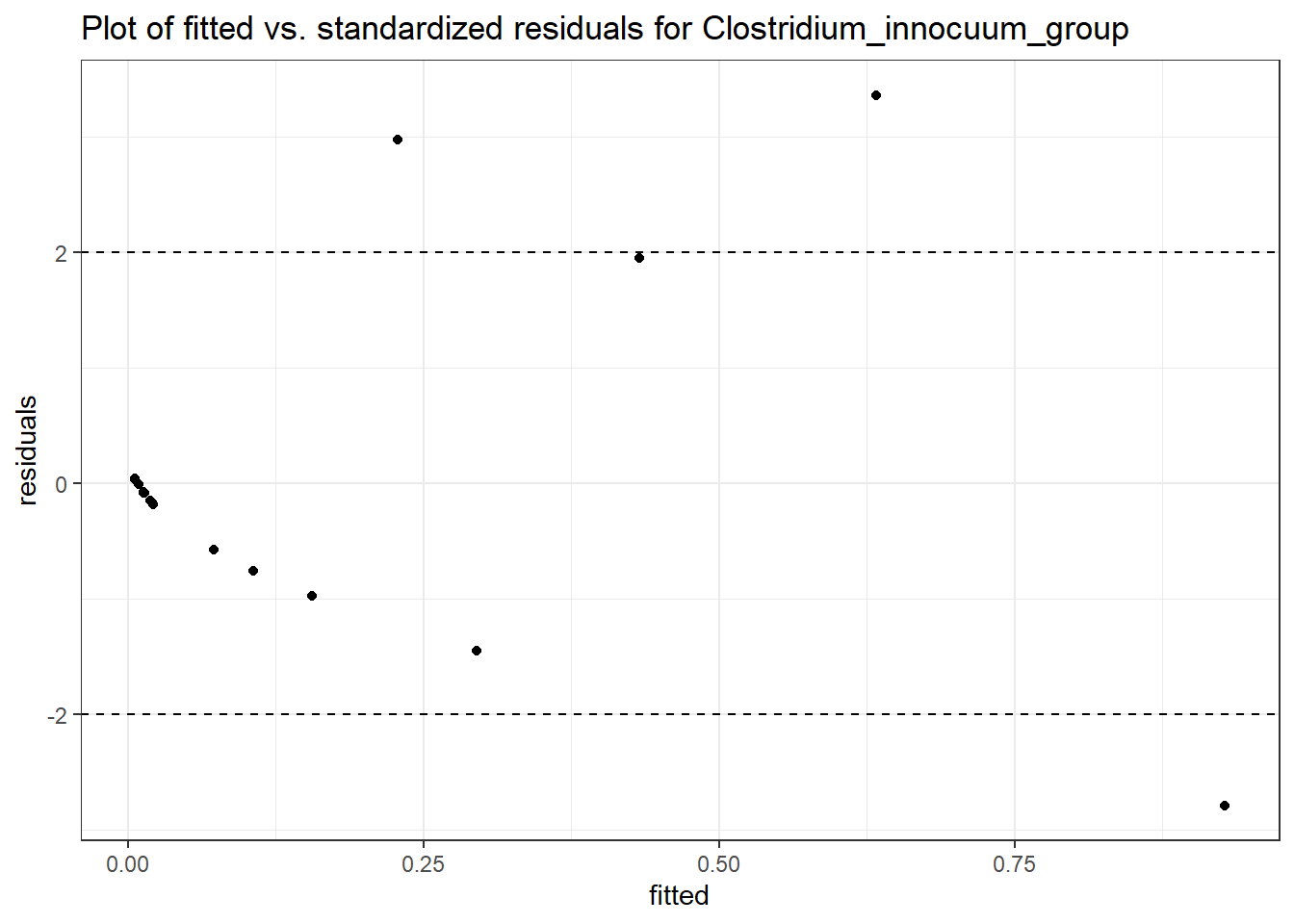
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_innocuum_group -3.71779 2.66710 -1.39395 0.16333
time Clostridium_innocuum_group -0.38200 0.48454 -0.78838 0.43048
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.2689
Significance test not available for random effects
#########################################
#########################################
Model: Eubacterium_brachy_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinom
Intraclass Correlation (ICC): 0.6579532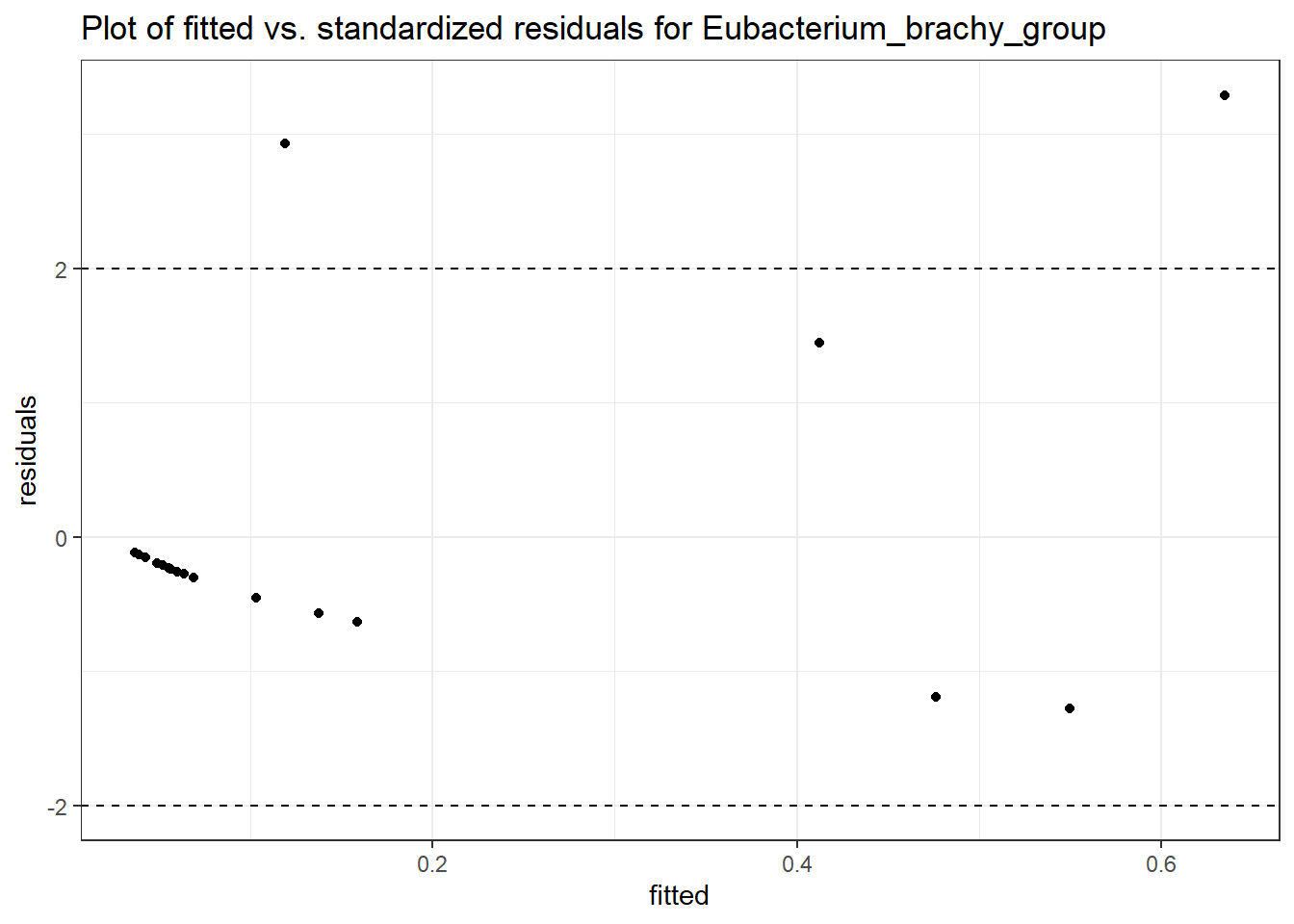
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Eubacterium_brachy_group -2.56655 1.43506 -1.78846 0.07370
time Eubacterium_brachy_group -0.14399 0.44157 -0.32609 0.74436
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.3869
Significance test not available for random effects
#########################################
#########################################
Model: Eubacterium_coprostanoligenes_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00785886 (tol = 0.002, component 1)
Intraclass Correlation (ICC): 0.7344367
Fixed Effects (change in log)
Outcome Estimate Std. Error z value
(Intercept) Eubacterium_coprostanoligenes_group 2.97333 0.60504 4.91428
time Eubacterium_coprostanoligenes_group 0.30149 0.18333 1.64452
Pr(>|z|)
(Intercept) 0.00000
time 0.10007
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.663
Significance test not available for random effects
#########################################
#########################################
Model: Eubacterium_eligens_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 1.030219e-09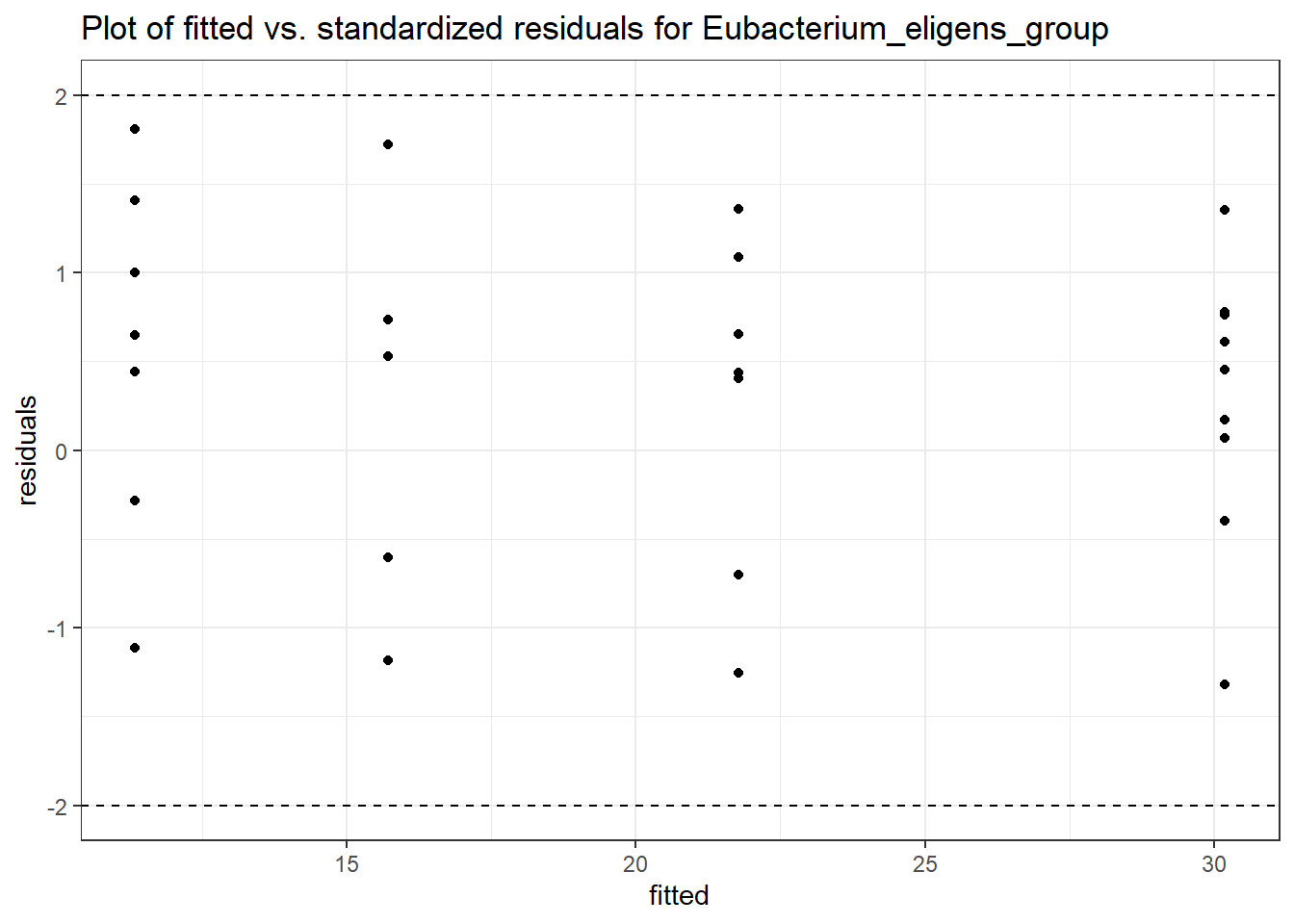

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Eubacterium_eligens_group 2.42791 0.44254 5.48633 0.00000
time Eubacterium_eligens_group 0.32656 0.24242 1.34712 0.17794
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.2097e-05
Significance test not available for random effects
#########################################
#########################################
Model: Eubacterium_hallii_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinom
Intraclass Correlation (ICC): 0.06215336
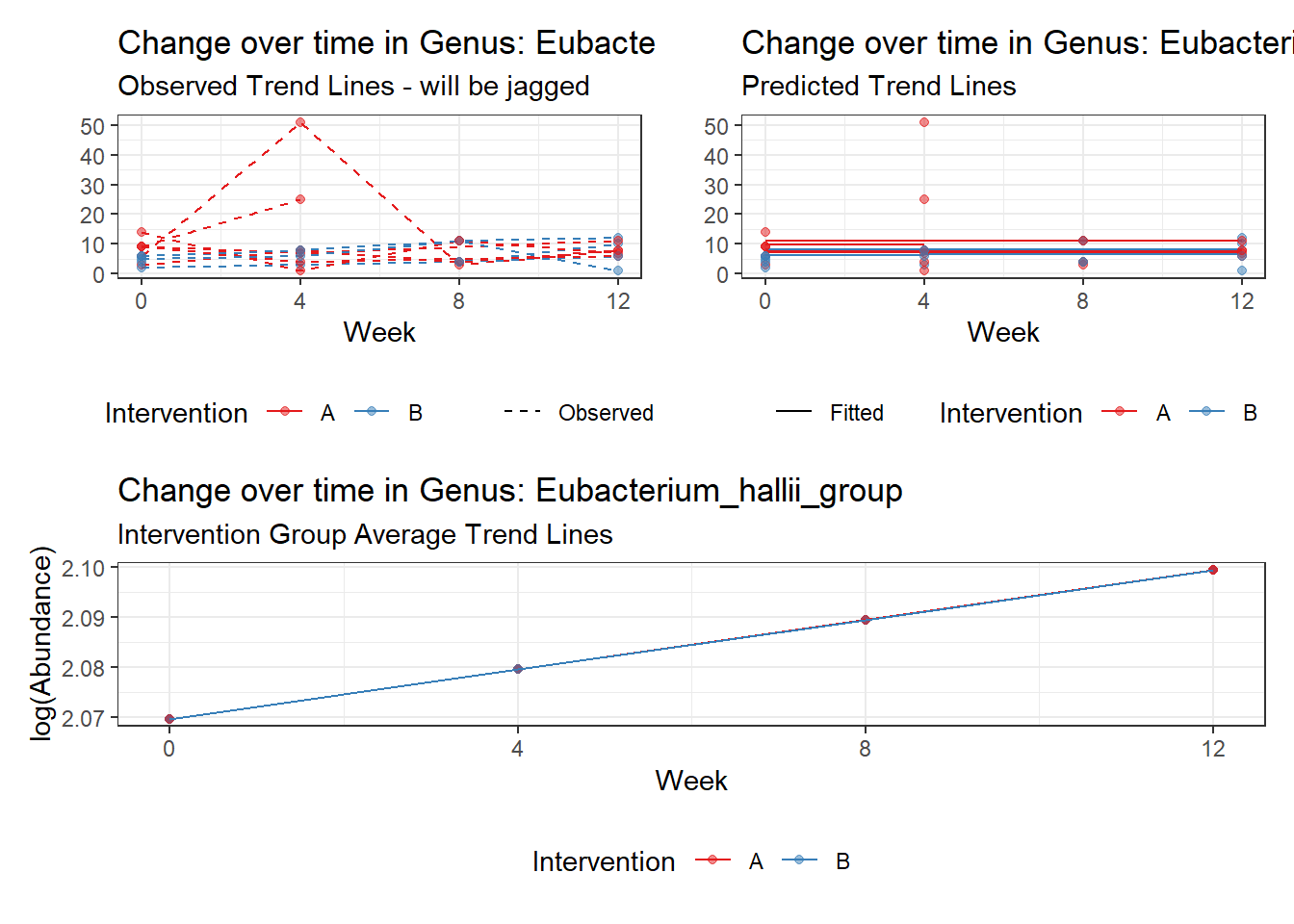
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Eubacterium_hallii_group 2.06967 0.20720 9.98883 0.0000
time Eubacterium_hallii_group 0.00989 0.10567 0.09364 0.9254
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.25743
Significance test not available for random effects
#########################################
#########################################
Model: Eubacterium_nodatum_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinom
Intraclass Correlation (ICC): 0.2060164
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Eubacterium_nodatum_group 0.43760 0.46465 0.94177 0.34631
time Eubacterium_nodatum_group 0.18645 0.26638 0.69995 0.48396
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.50938
Significance test not available for random effects
#########################################
#########################################
Model: Eubacterium_ventriosum_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinomboundary (singular) fit: see ?isSingular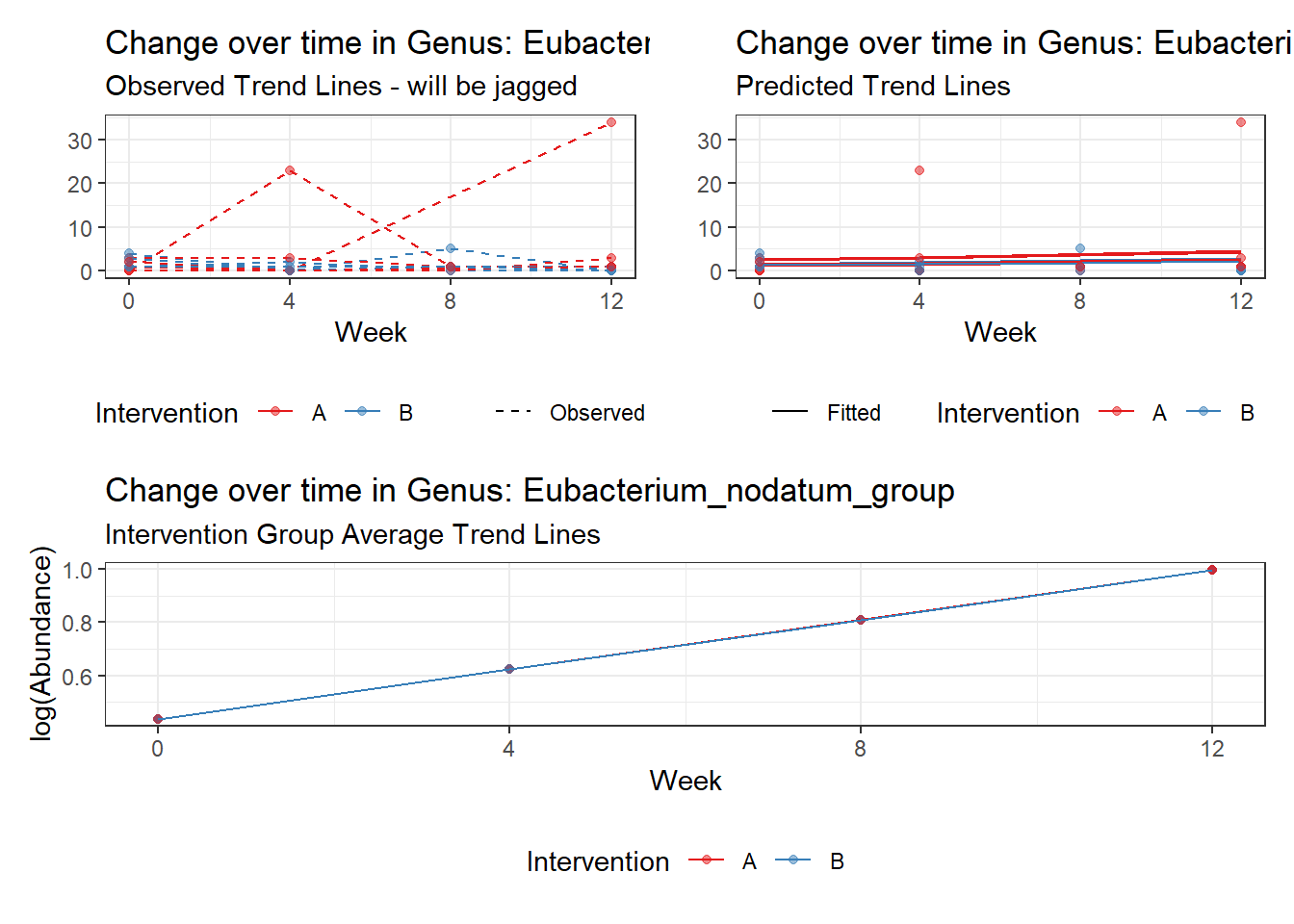
Intraclass Correlation (ICC): 7.192072e-10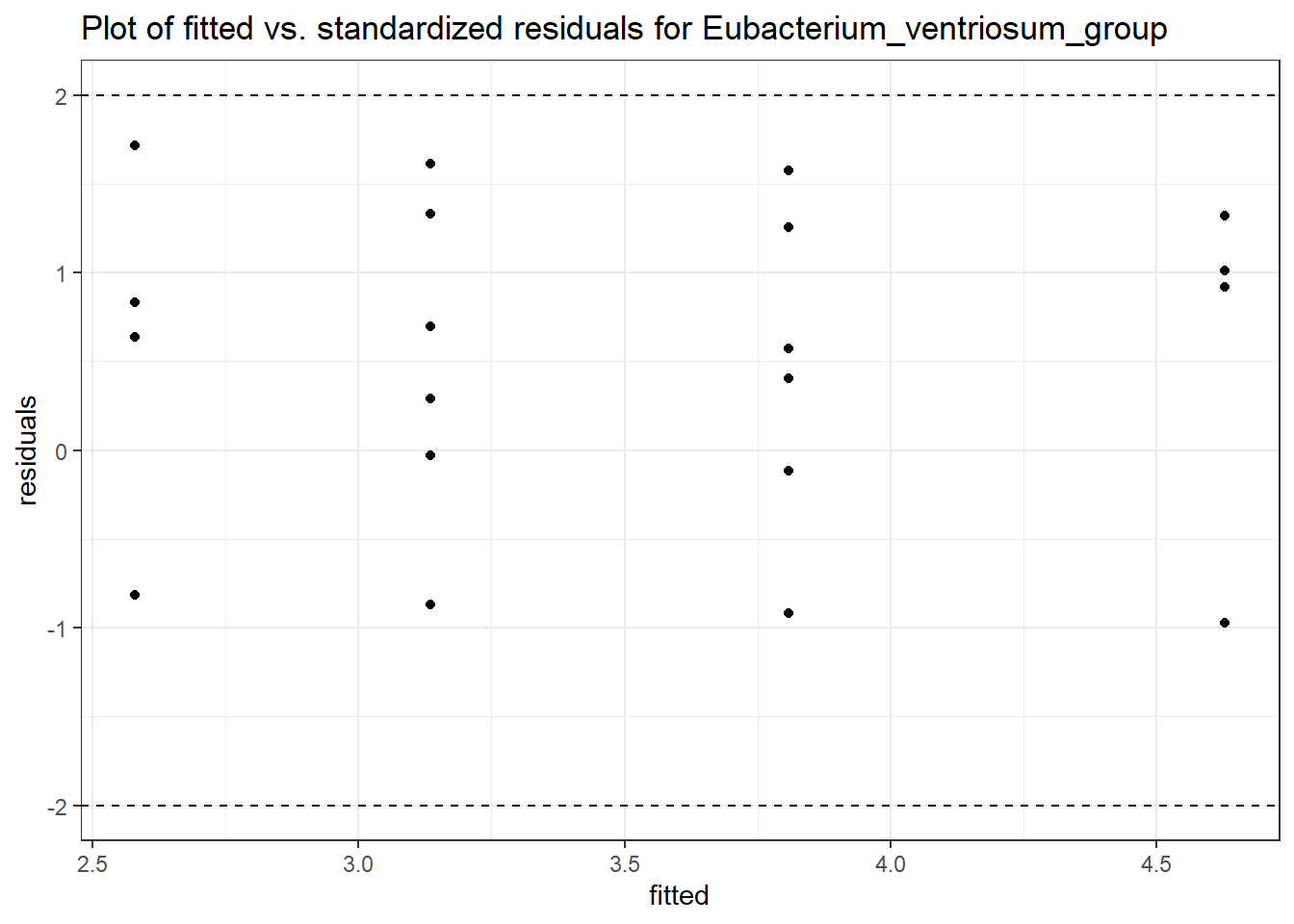
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Eubacterium_ventriosum_group 0.94761 0.49693 1.90692 0.05653
time Eubacterium_ventriosum_group 0.19494 0.27533 0.70801 0.47894
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6818e-05
Significance test not available for random effects
#########################################
#########################################
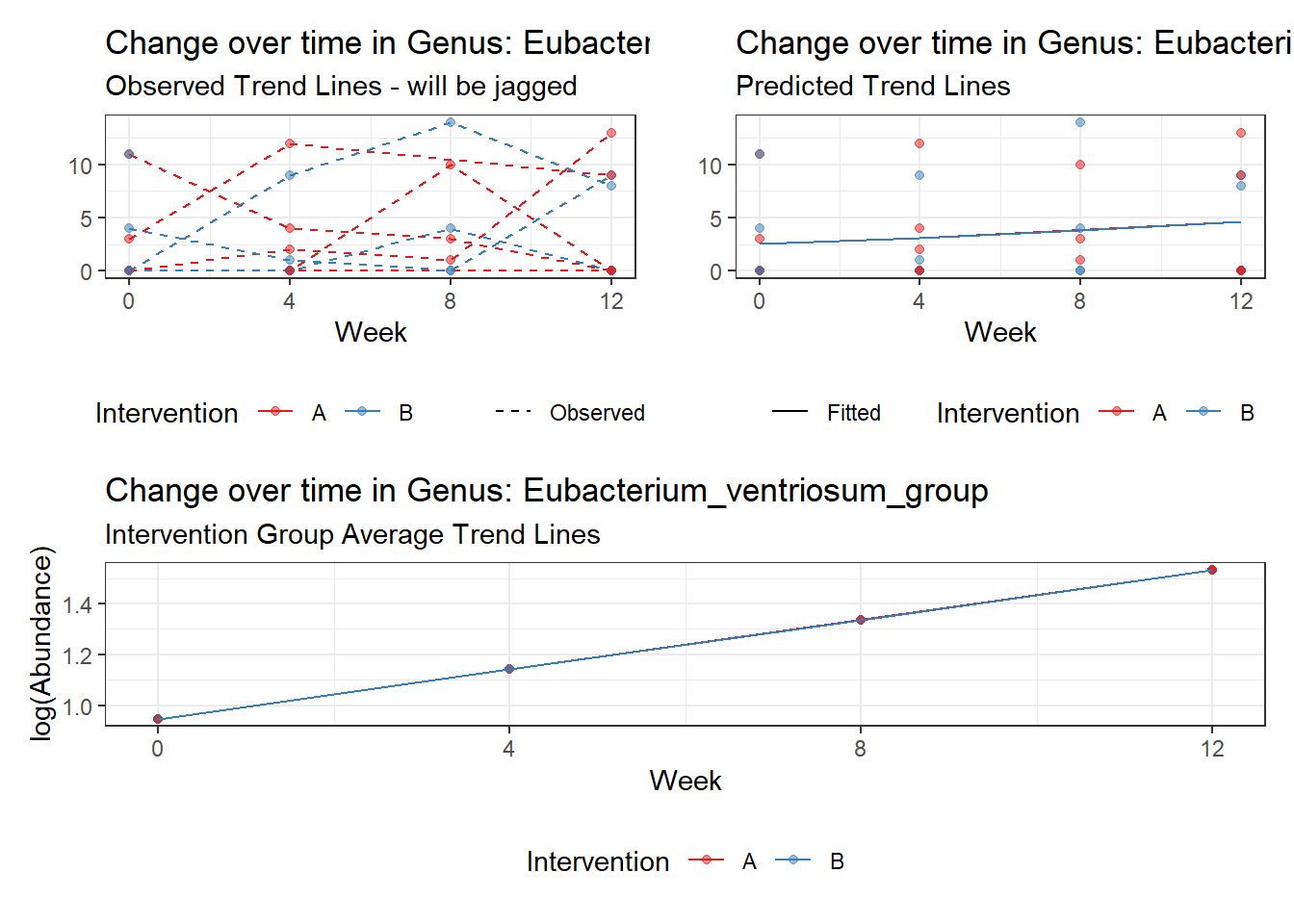
Model: Eubacterium_xylanophilum_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinom
Intraclass Correlation (ICC): 0.8379412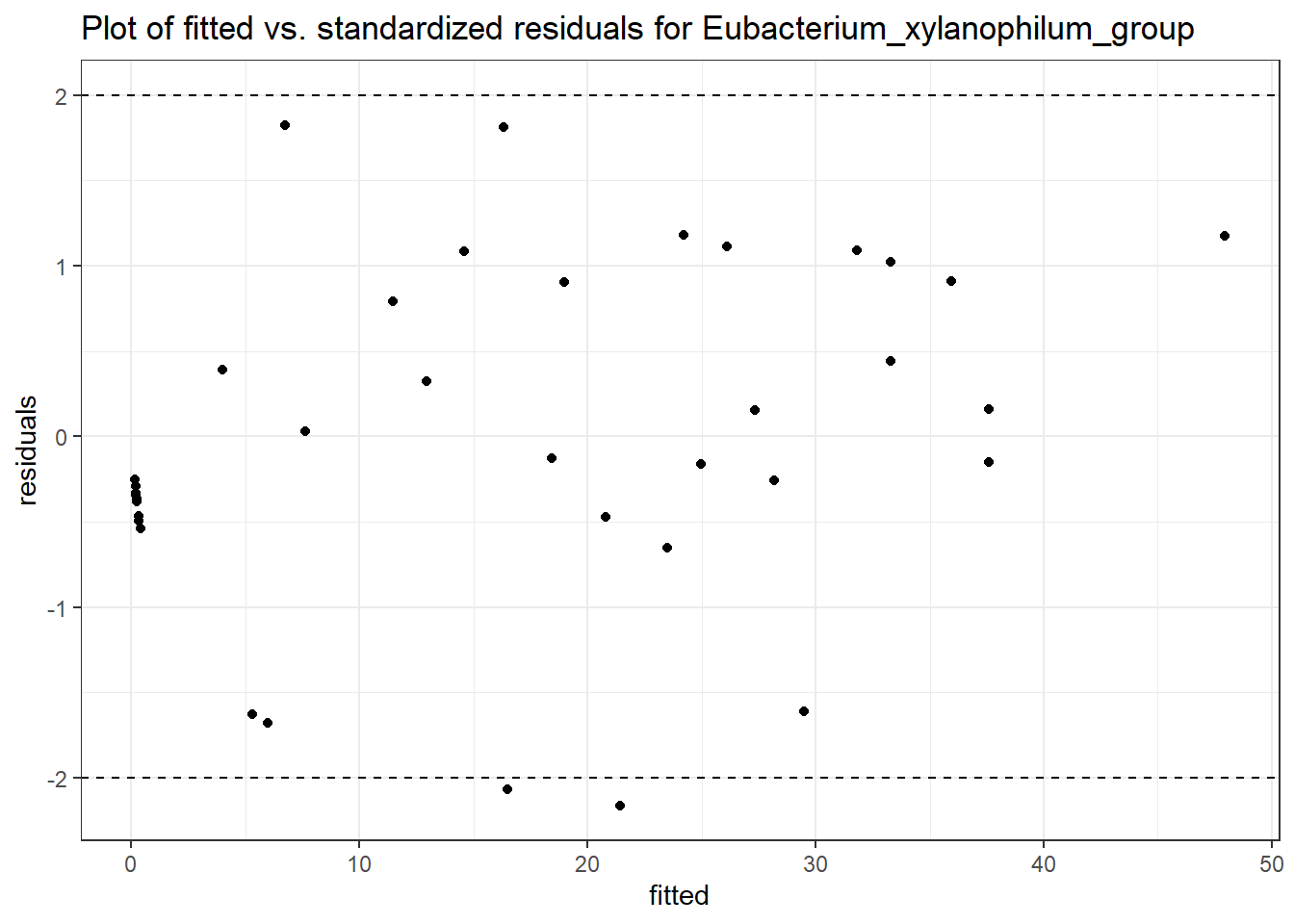
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Eubacterium_xylanophilum_group 1.40195 0.84049 1.66802 0.09531
time Eubacterium_xylanophilum_group 0.12155 0.21379 0.56853 0.56968
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.2739
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_gauvreauii_group ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000421de8c0>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
unable to evaluate scaled gradientWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Hessian is numerically singular: parameters are not uniquely determinedWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Hessian is numerically singular: parameters are not uniquely determinedError in factory(refitNB, types = c("message", "warning"))(lastfit, theta = exp(t), : pwrssUpdate did not converge in (maxit) iterations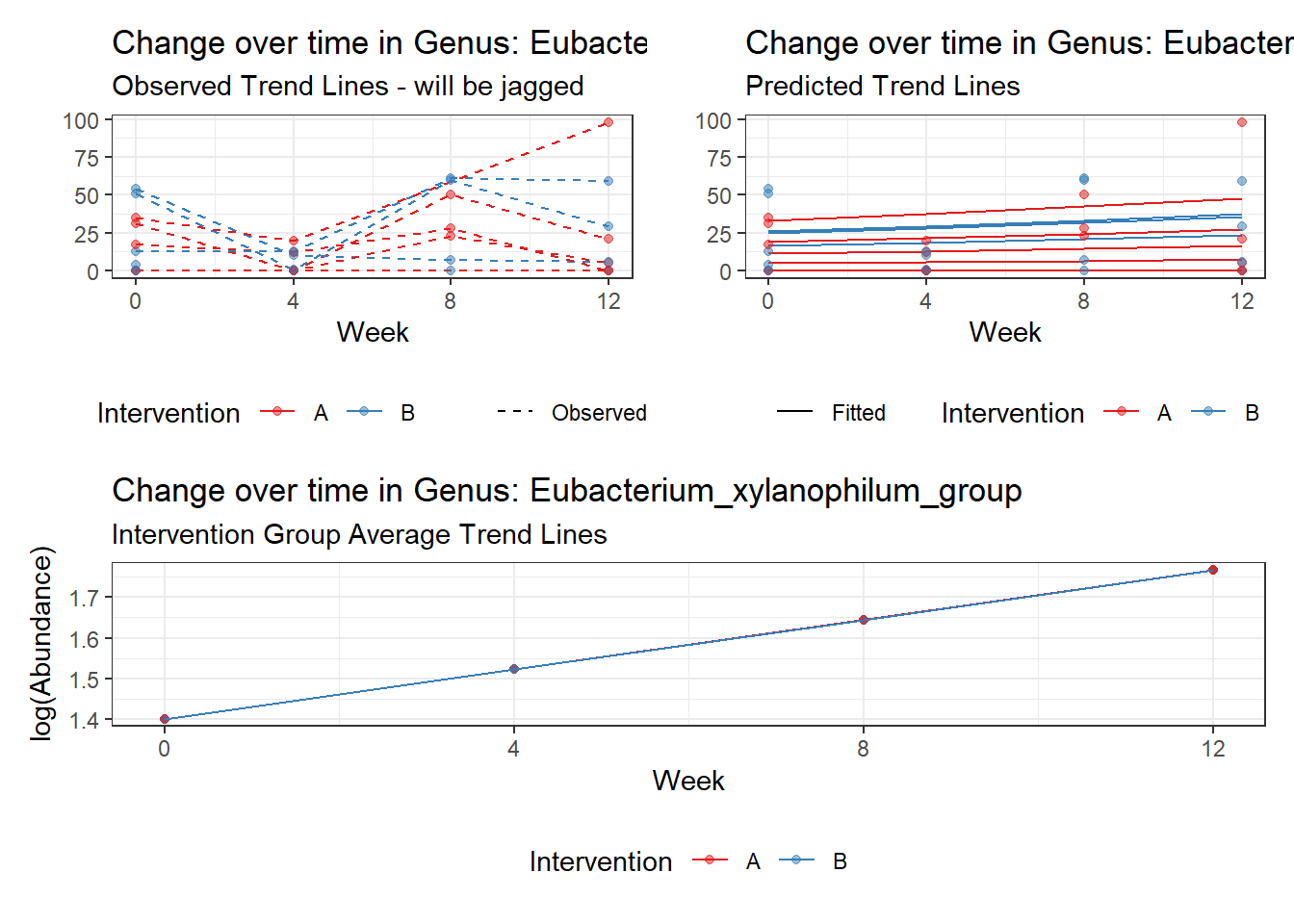
So, we found that time does significantly influence at least some of the bacteria over time. This is contrary to the results when we used the normal theory model.
Covariates: Time + Gender
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Gender)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
Gender is coded as female (effect of being female compared to males).
genus.fit2 <- glmm_microbiome(mydata=microbiome_data,model.number=2,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + time + female + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.827486

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 2.90035 1.12841 2.57031 0.01016
time Akkermansia 0.00500 0.00920 0.54291 0.58719
female Akkermansia 2.50316 1.39938 1.78876 0.07365
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1901
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.1888678

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 8.19179 0.24135 33.94213 0.00000
time Bacteroides -0.05158 0.00312 -16.54937 0.00000
female Bacteroides -0.83181 0.30258 -2.74903 0.00598
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.48254
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.5644021
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.42125 0.58018 4.17327 0.00003
time Bifidobacterium 0.04989 0.03554 1.40394 0.16034
female Bifidobacterium -0.31708 0.72595 -0.43679 0.66227
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.1383
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.6516701
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 -0.30728 0.81914 -0.37513 0.70756
time Clostridium_sensu_stricto_1 0.23572 0.06296 3.74375 0.00018
female Clostridium_sensu_stricto_1 0.95734 0.97098 0.98595 0.32416
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.3678
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.1279272
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.11194 0.20609 15.09987 0.00000
time Dorea -0.17318 0.02856 -6.06273 0.00000
female Dorea 0.45727 0.25358 1.80324 0.07135
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.38301
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.3238632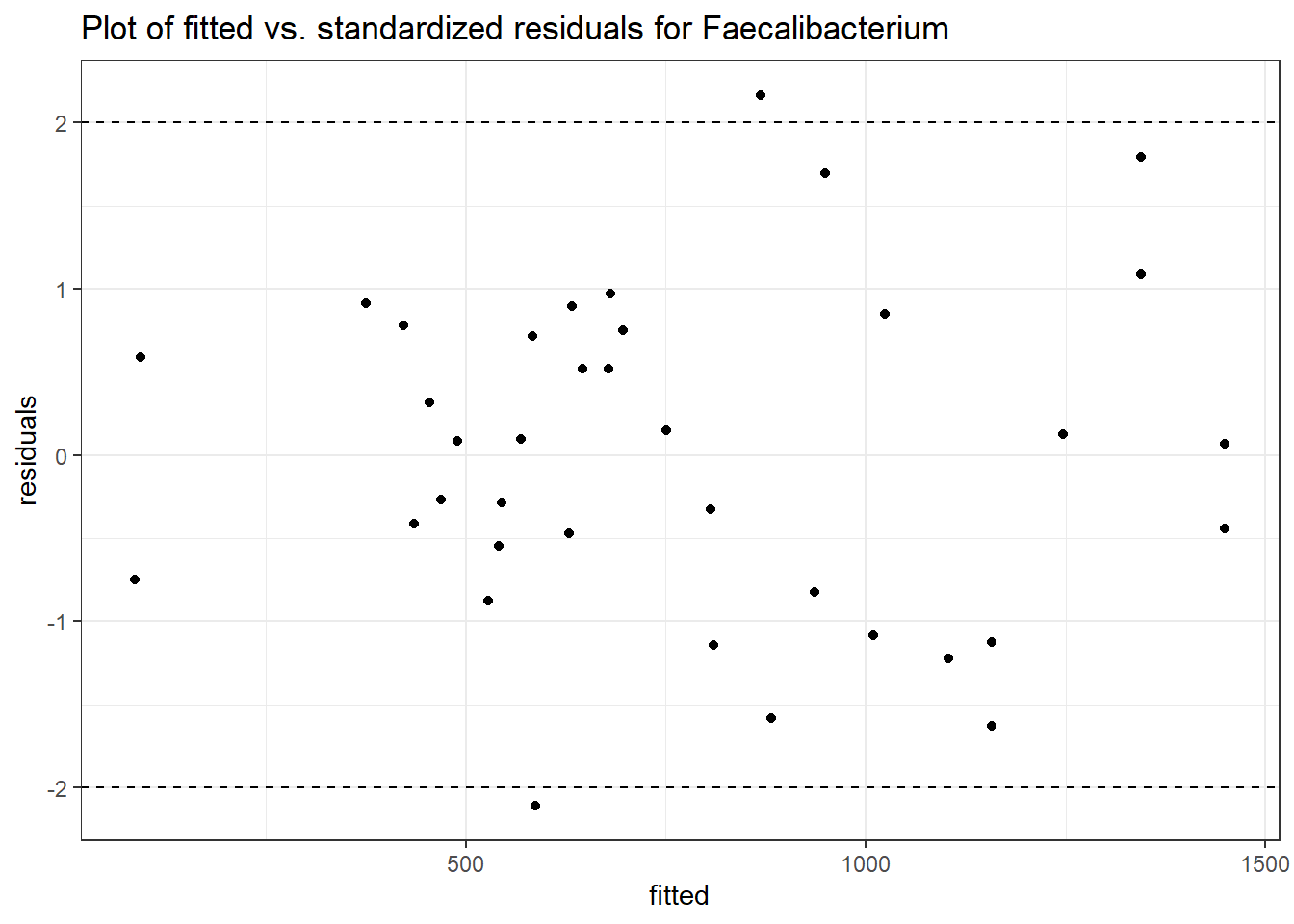
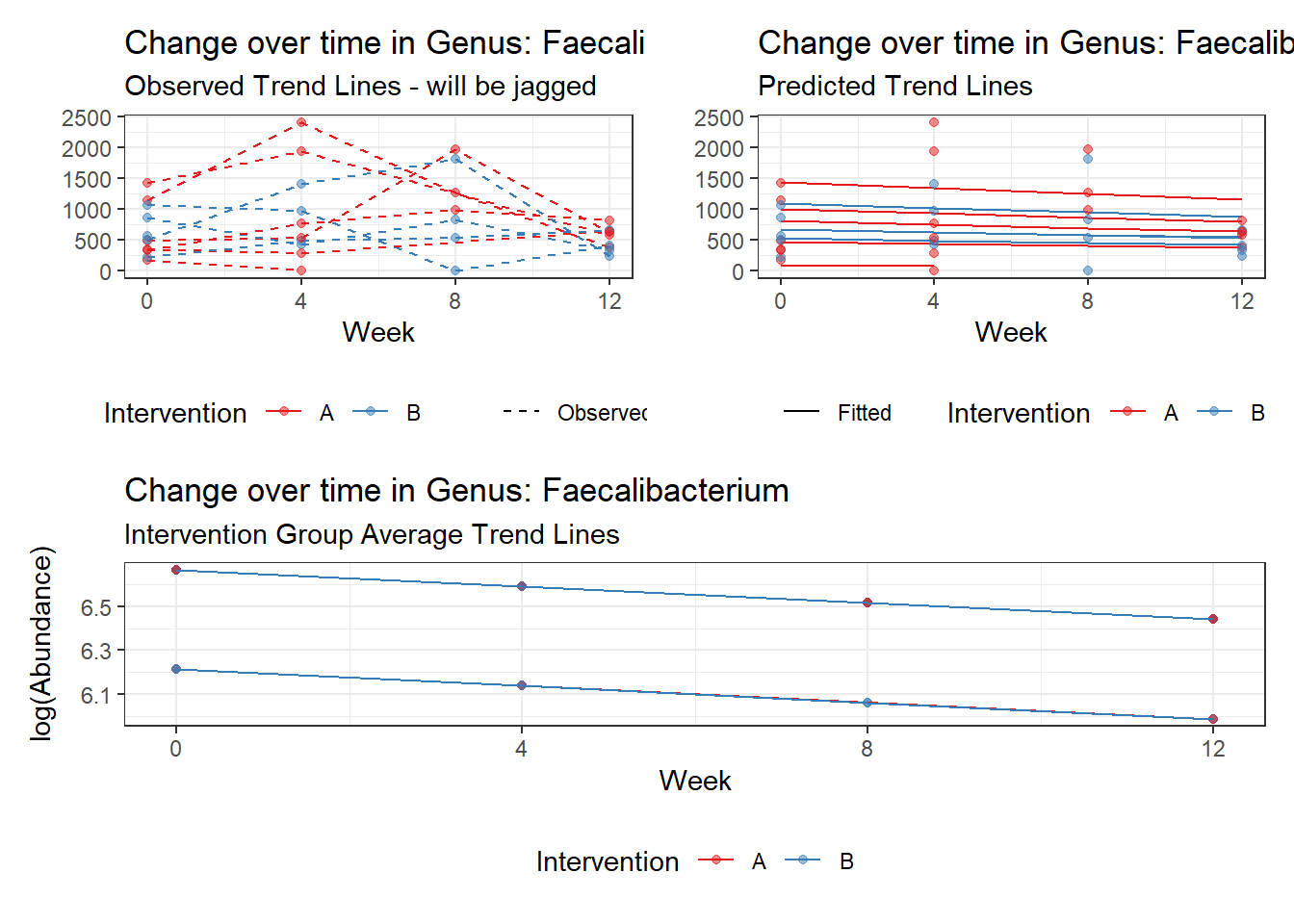
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.21180 0.34670 17.91693 0.00000
time Faecalibacterium -0.07486 0.00529 -14.15940 0.00000
female Faecalibacterium 0.45352 0.43437 1.04409 0.29644
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.69209
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.2531228
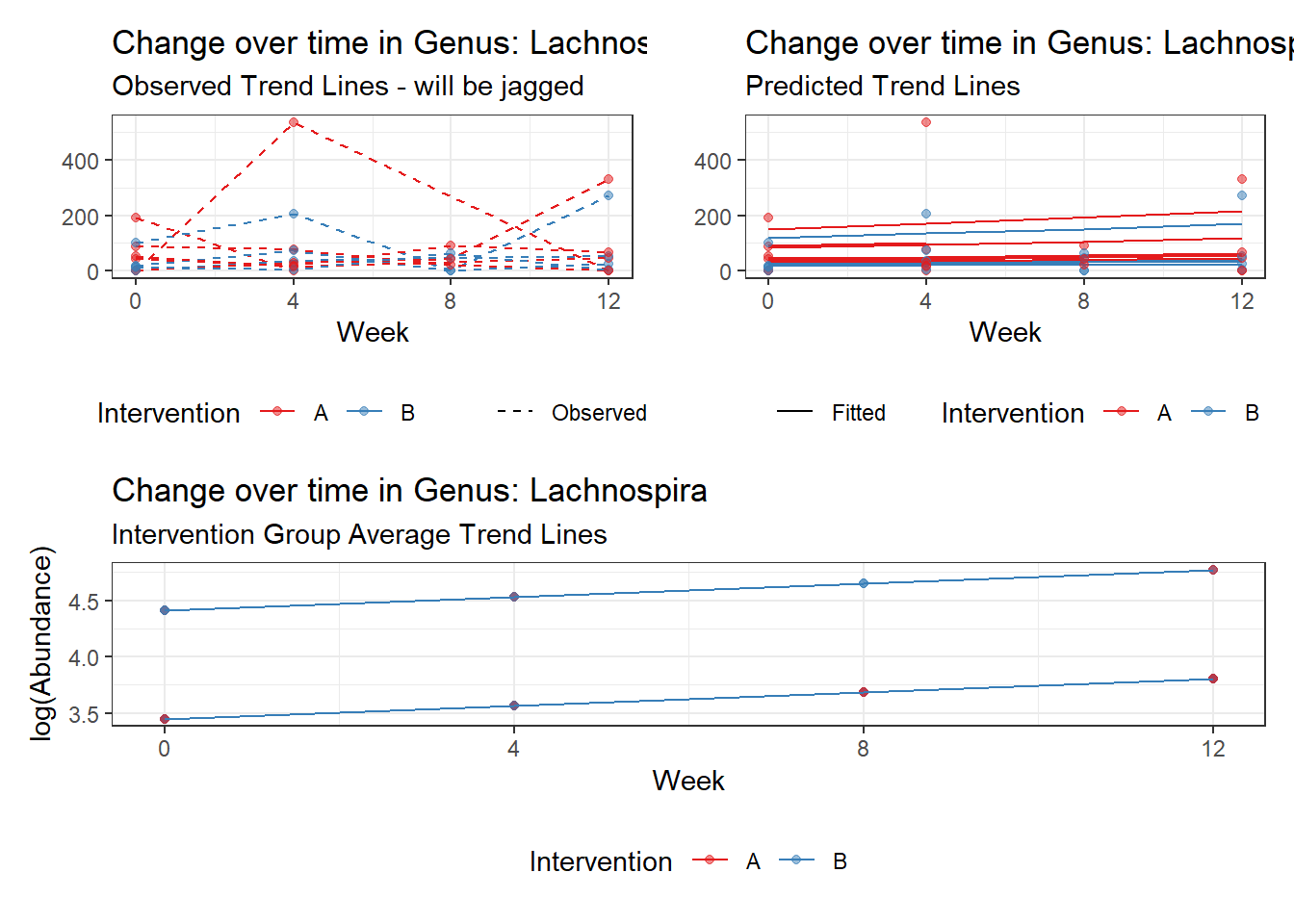
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.41293 0.29373 15.02358 0.00000
time Lachnospira 0.11902 0.01758 6.77066 0.00000
female Lachnospira -0.96329 0.36899 -2.61058 0.00904
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.58216
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.8488898
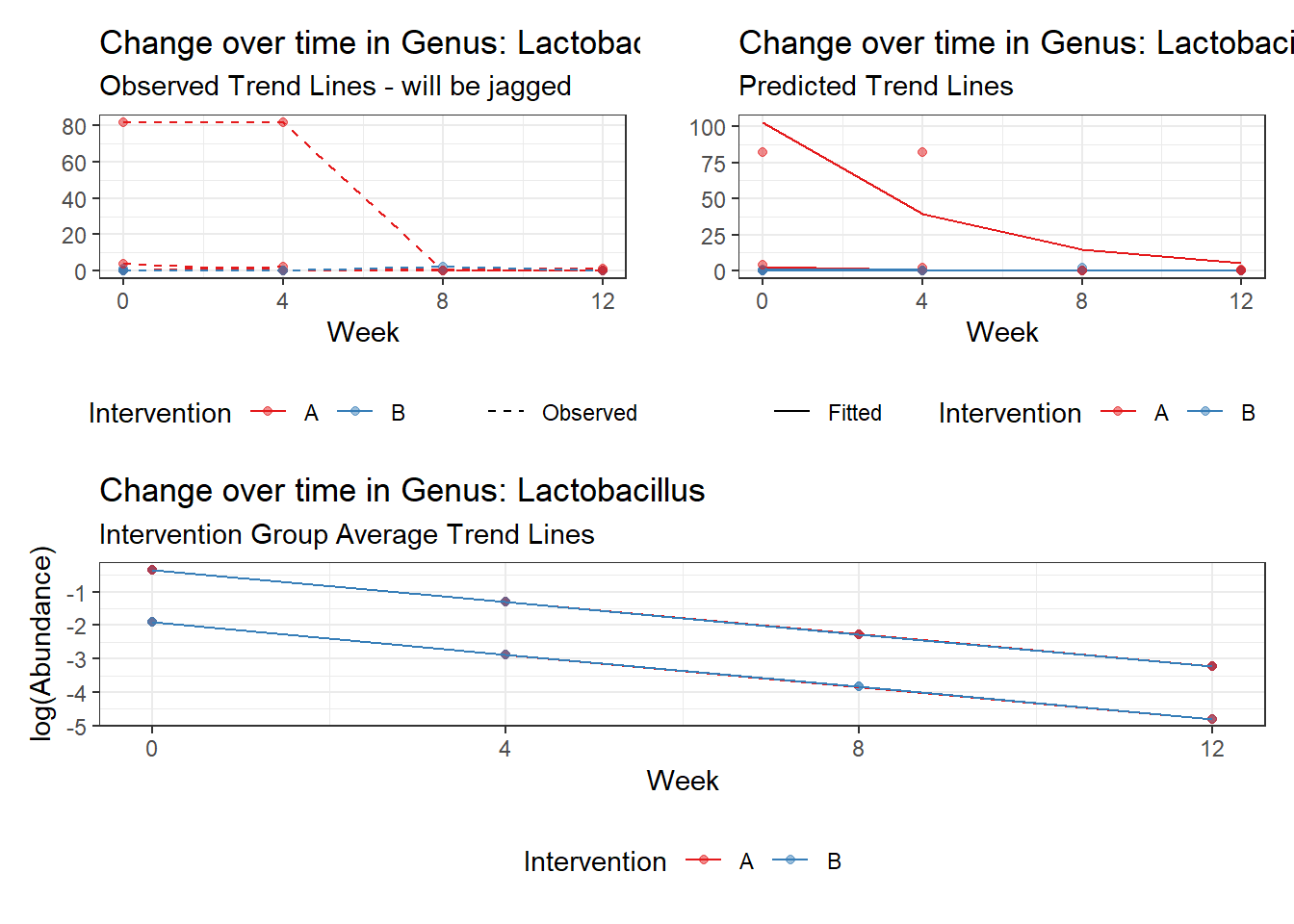
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus -1.91335 1.54682 -1.23696 0.21610
time Lactobacillus -0.96010 0.09523 -10.08183 0.00000
female Lactobacillus 1.56326 1.79150 0.87260 0.38288
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3702
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.7577428
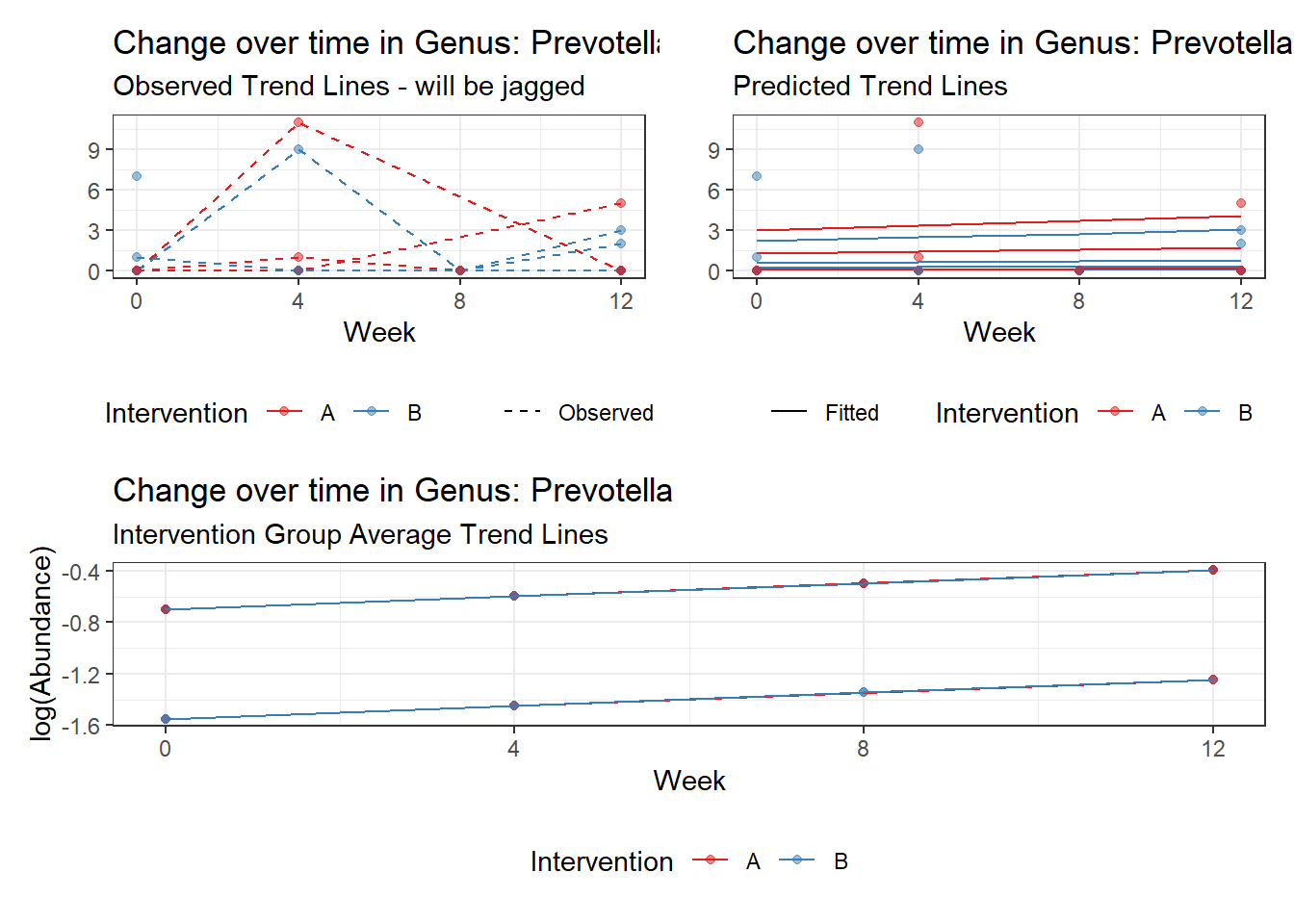
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.54861 1.15302 -1.34310 0.17924
time Prevotella 0.10204 0.14714 0.69346 0.48802
female Prevotella 0.84819 1.30493 0.64999 0.51570
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7686
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.700194

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 0.88336 0.86754 1.01823 0.30857
time Roseburia 0.09582 0.03045 3.14675 0.00165
female Roseburia 2.01509 1.04255 1.93285 0.05326
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.5282
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046a3d810>
Link: poisson
Intraclass Correlation (ICC): 0.516513
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 1.12440 0.56645 1.98500 0.04714
time Ruminococcus_1 0.25713 0.01598 16.08648 0.00000
female Ruminococcus_1 3.17454 0.68811 4.61340 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0336
Significance test not available for random effectsgenus.fit2b <- glmm_microbiome(mydata=microbiome_data,model.number=2,
taxa.level="Genus", taxa.subset = backNames,
link="negbinom",
model="1 + time + female + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced
Intraclass Correlation (ICC): 0.8291102
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 2.88130 1.11034 2.59496 0.00946
time Akkermansia 0.00202 0.00937 0.21506 0.82972
female Akkermansia 2.52597 1.37724 1.83408 0.06664
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.2027
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinom
Intraclass Correlation (ICC): 0.133748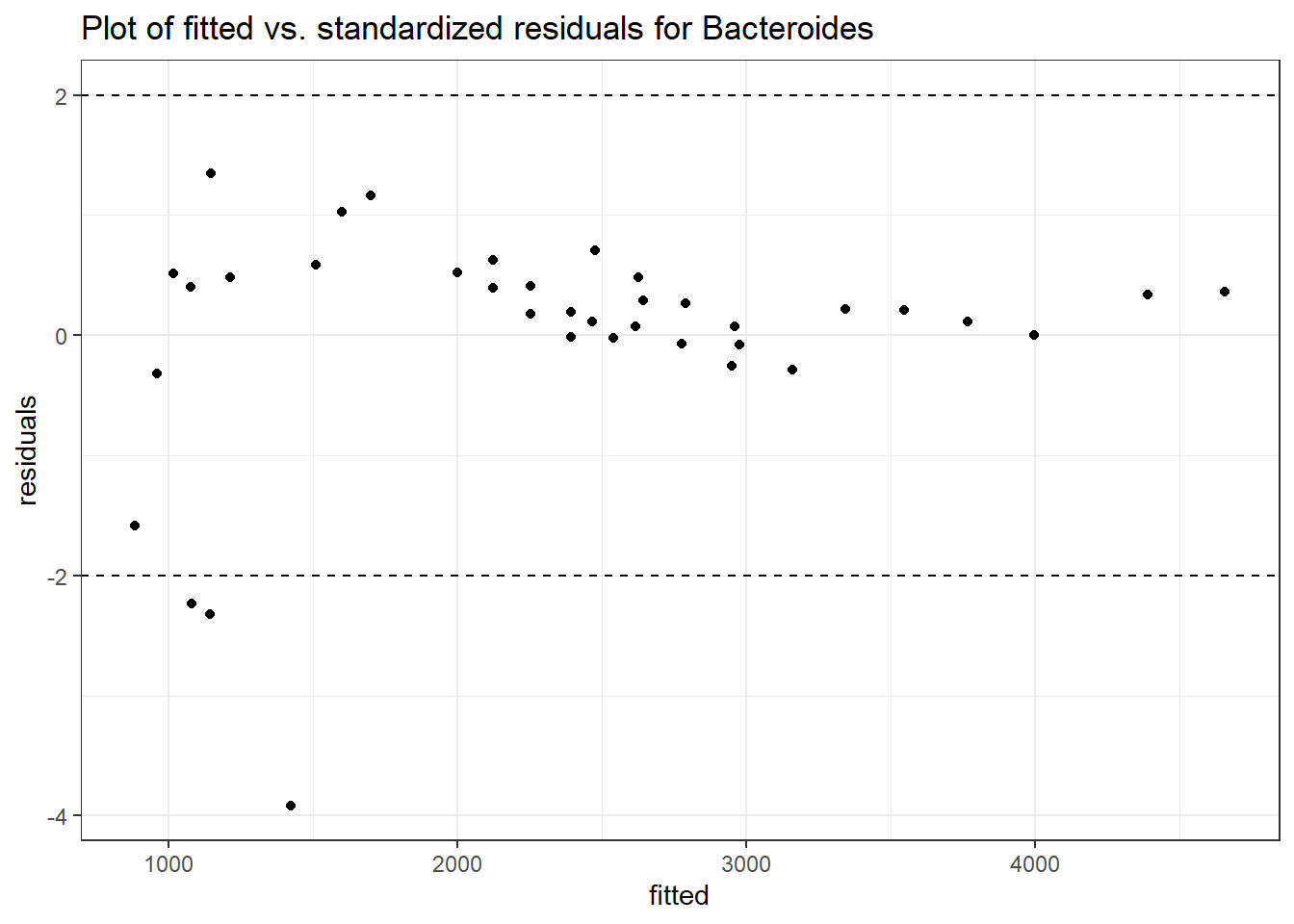
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 8.19642 0.22213 36.89872 0.00000
time Bacteroides -0.05947 0.04506 -1.31991 0.18687
female Bacteroides -0.77035 0.27213 -2.83085 0.00464
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.39294
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinom
Intraclass Correlation (ICC): 0.3254106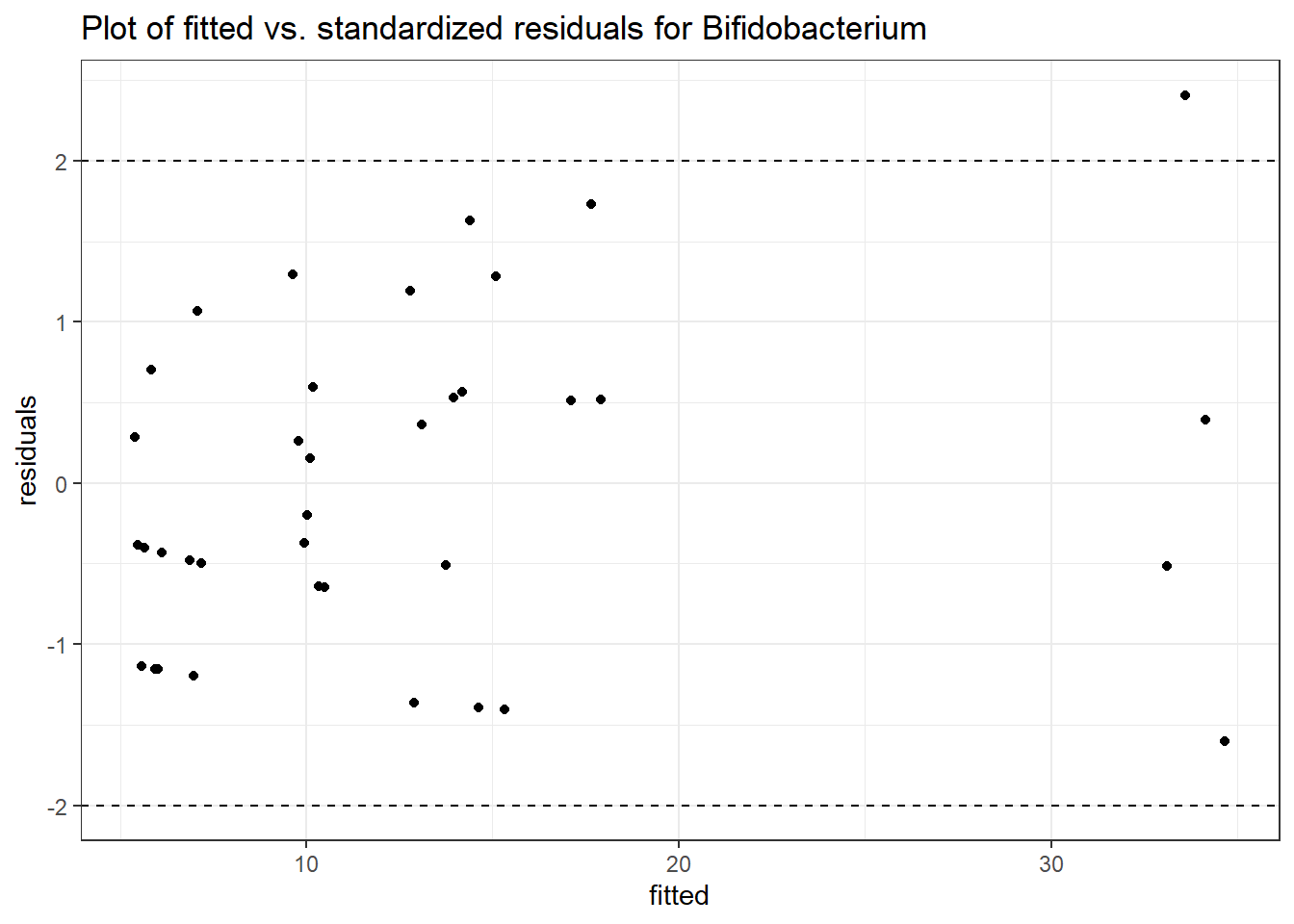
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.79803 0.80842 3.46110 0.00054
time Bifidobacterium -0.01545 0.28210 -0.05476 0.95633
female Bifidobacterium -0.52522 0.74936 -0.70090 0.48337
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.69454
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinom
Intraclass Correlation (ICC): 0.4701544
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.48752 1.14457 0.42594 0.67015
time Clostridium_sensu_stricto_1 0.10443 0.38421 0.27181 0.78577
female Clostridium_sensu_stricto_1 0.57813 1.14808 0.50357 0.61457
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.94199
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinom
Intraclass Correlation (ICC): 0.0162737
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.17062 0.25033 12.66567 0.00000
time Dorea -0.12826 0.09574 -1.33970 0.18034
female Dorea 0.44061 0.25913 1.70030 0.08907
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.12862
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinomboundary (singular) fit: see ?isSingular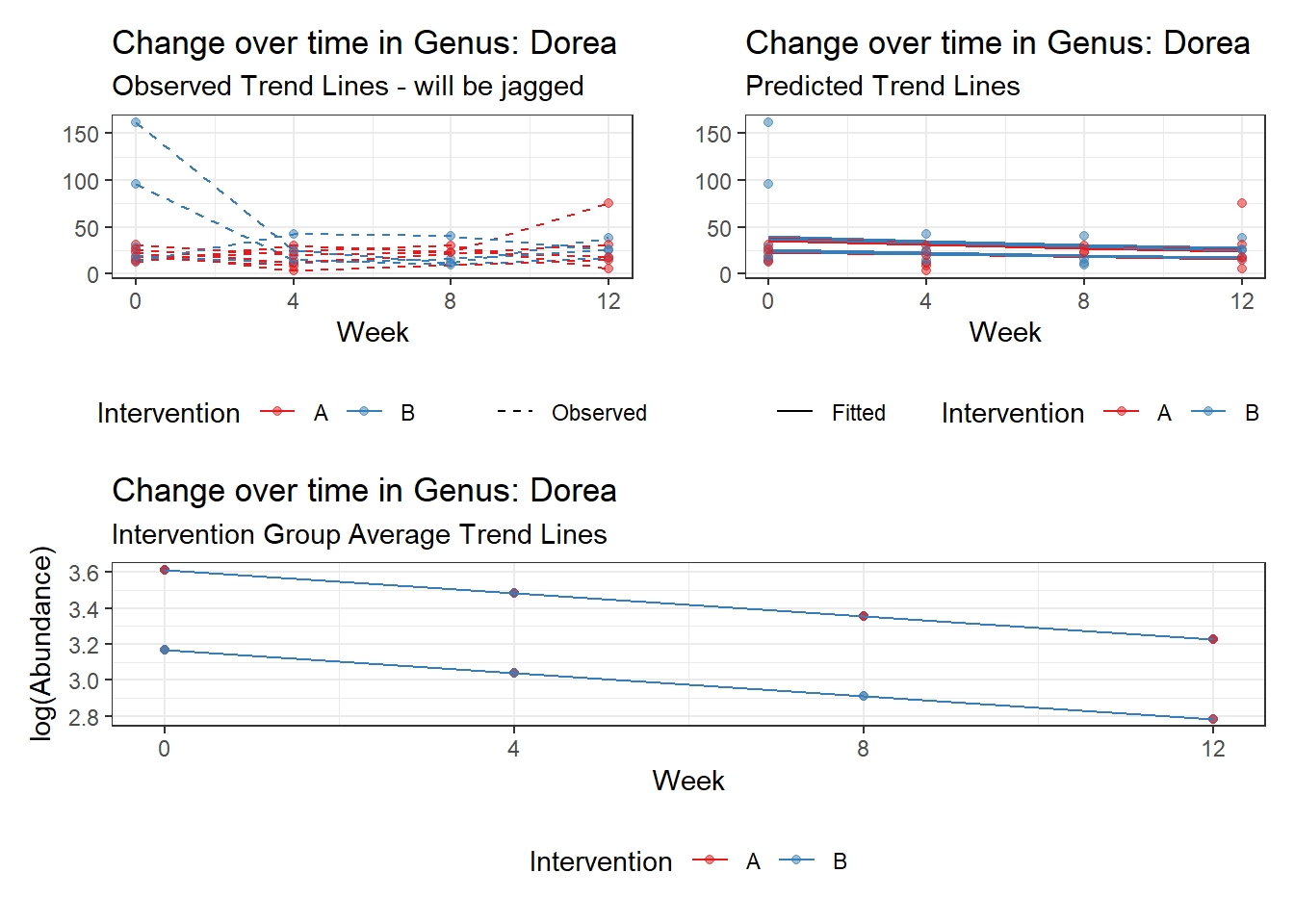
Intraclass Correlation (ICC): 3.004096e-12
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.59938 0.31396 21.01948 0.00000
time Faecalibacterium -0.05711 0.15682 -0.36416 0.71574
female Faecalibacterium 0.18376 0.34038 0.53987 0.58929
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7332e-06
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reached
Warning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, : NaNs
produced
Intraclass Correlation (ICC): 0.2529383
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.41208 0.29381 15.01699 0.000
time Lachnospira 0.12010 0.01779 6.75034 0.000
female Lachnospira -0.96411 0.36911 -2.61202 0.009
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.58187
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00404453 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 1.559771e-10
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus 0.45126 2.45796 0.18359 0.85434
time Lactobacillus -1.02718 0.95942 -1.07062 0.28434
female Lactobacillus 2.19788 2.01763 1.08934 0.27600
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.2489e-05
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003f8a4ed8>
Link: negbinomError in pwrssUpdate(pp, resp, tol = tolPwrss, GQmat = GHrule(0L), compDev = compDev, : pwrssUpdate did not converge in (maxit) iterations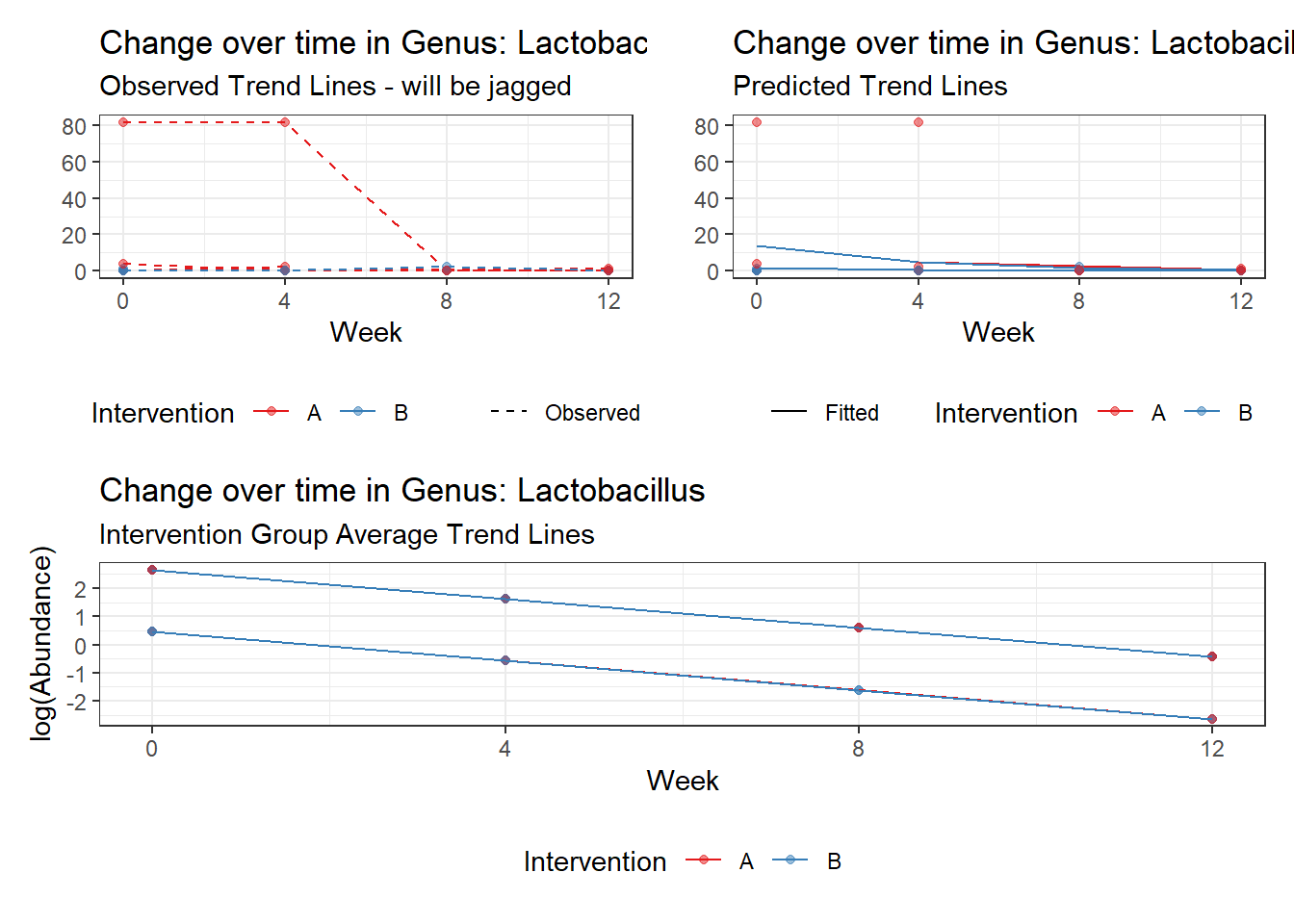
Controlling for the effect of time, Gender was only signficant for Bacteroidetes and Firmicutes. We will take out gender out of model to test other effects, but will come back when testing intervention.
Covariates: Time + Ethnicity
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Ethnicity)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
Ethnicity is coded as Hispanic (effect of being Hispanic compared to White, Asian or Native American). Groups were coded this way due to sample size.
genus.fit3 <- glmm_microbiome(mydata=microbiome_data,model.number=3,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.8512603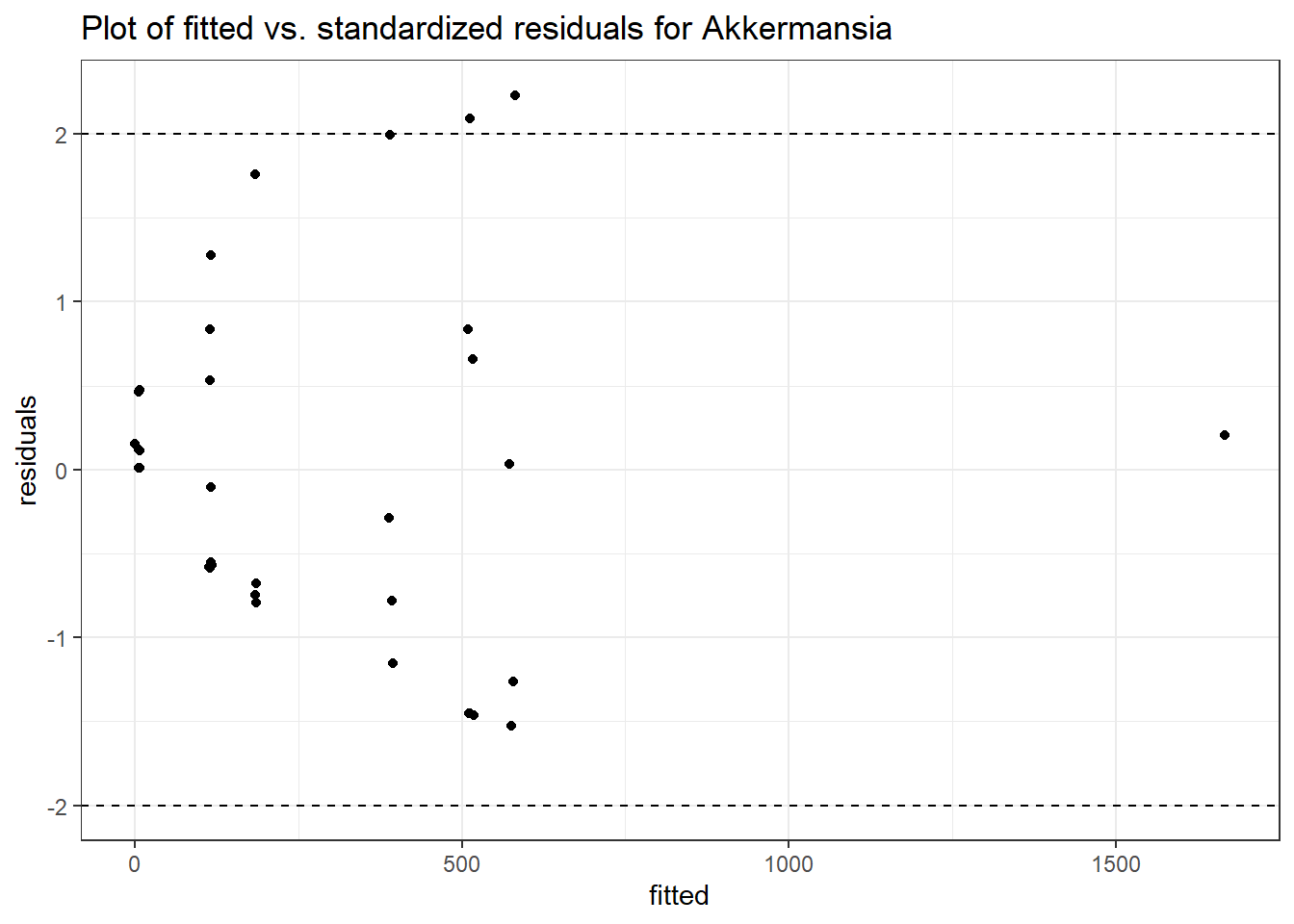
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 3.81822 1.19875 3.18516 0.00145
time Akkermansia 0.00501 0.00920 0.54458 0.58604
hispanic Akkermansia 1.09275 1.50857 0.72436 0.46884
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3923
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.2316701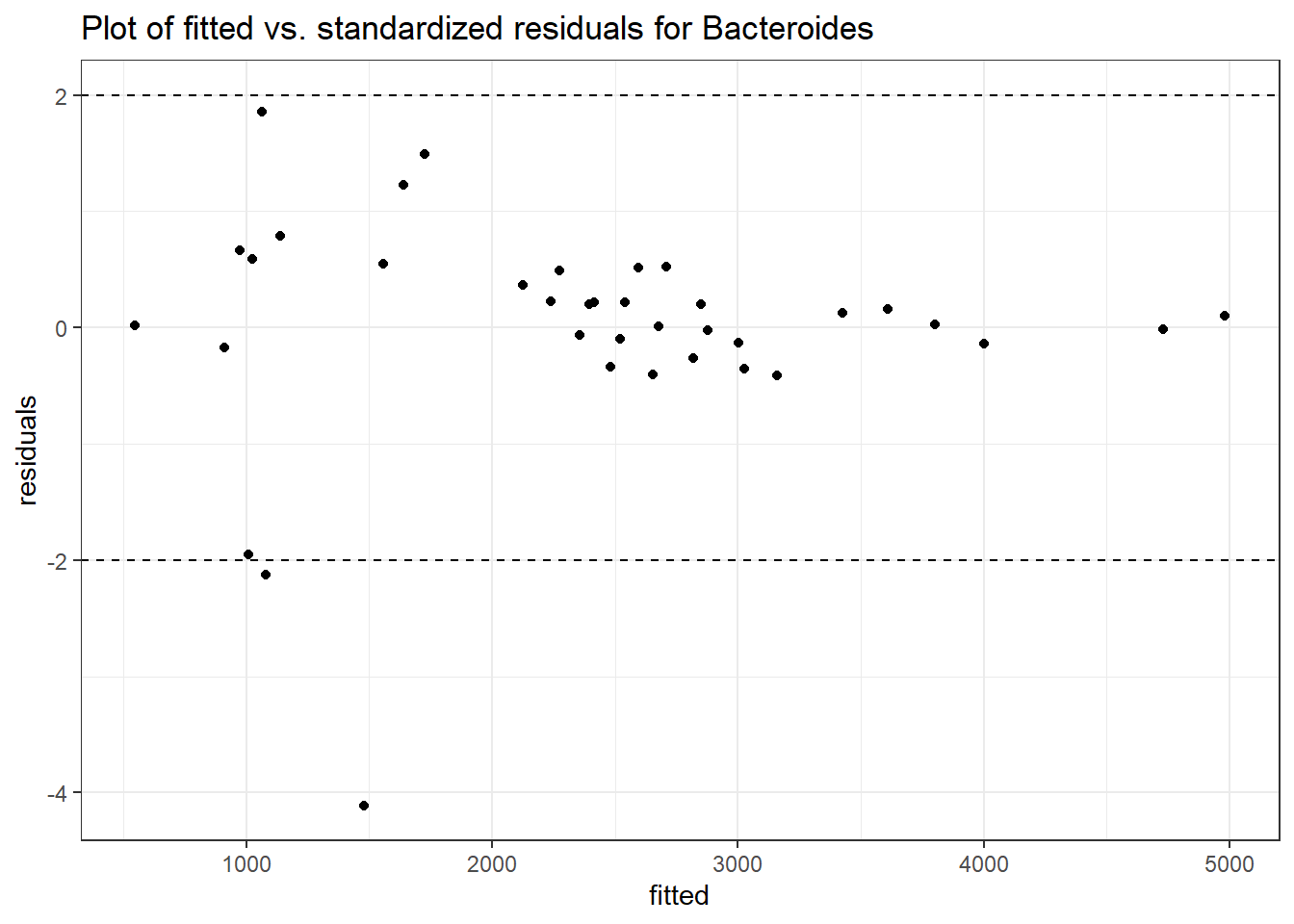

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 8.06317 0.27463 29.35963 0.00000
time Bacteroides -0.05161 0.00312 -16.56012 0.00000
hispanic Bacteroides -0.62982 0.34430 -1.82929 0.06736
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.54911
Significance test not available for random effects
#########################################
#########################################
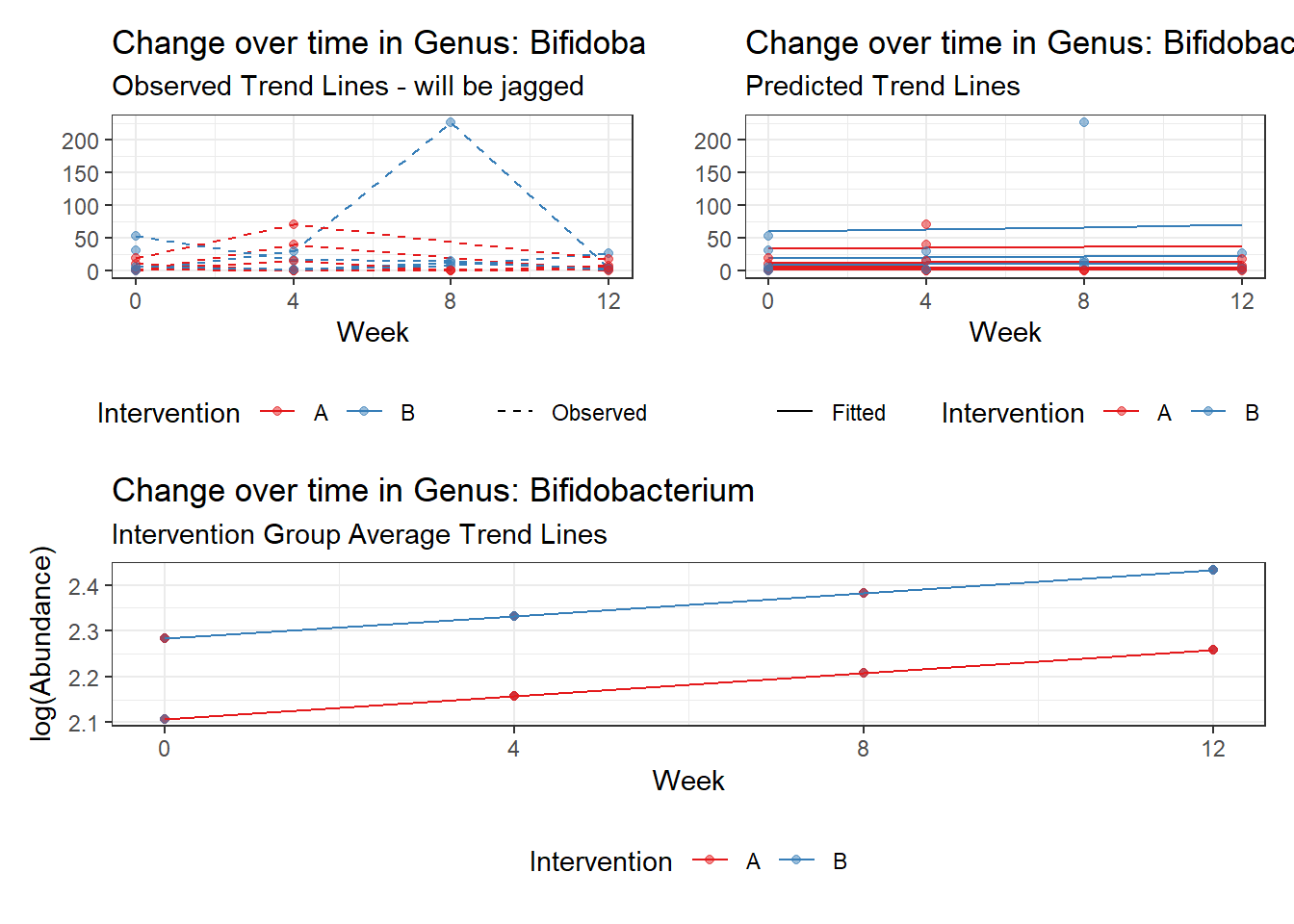
Model: Bifidobacterium ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.5675625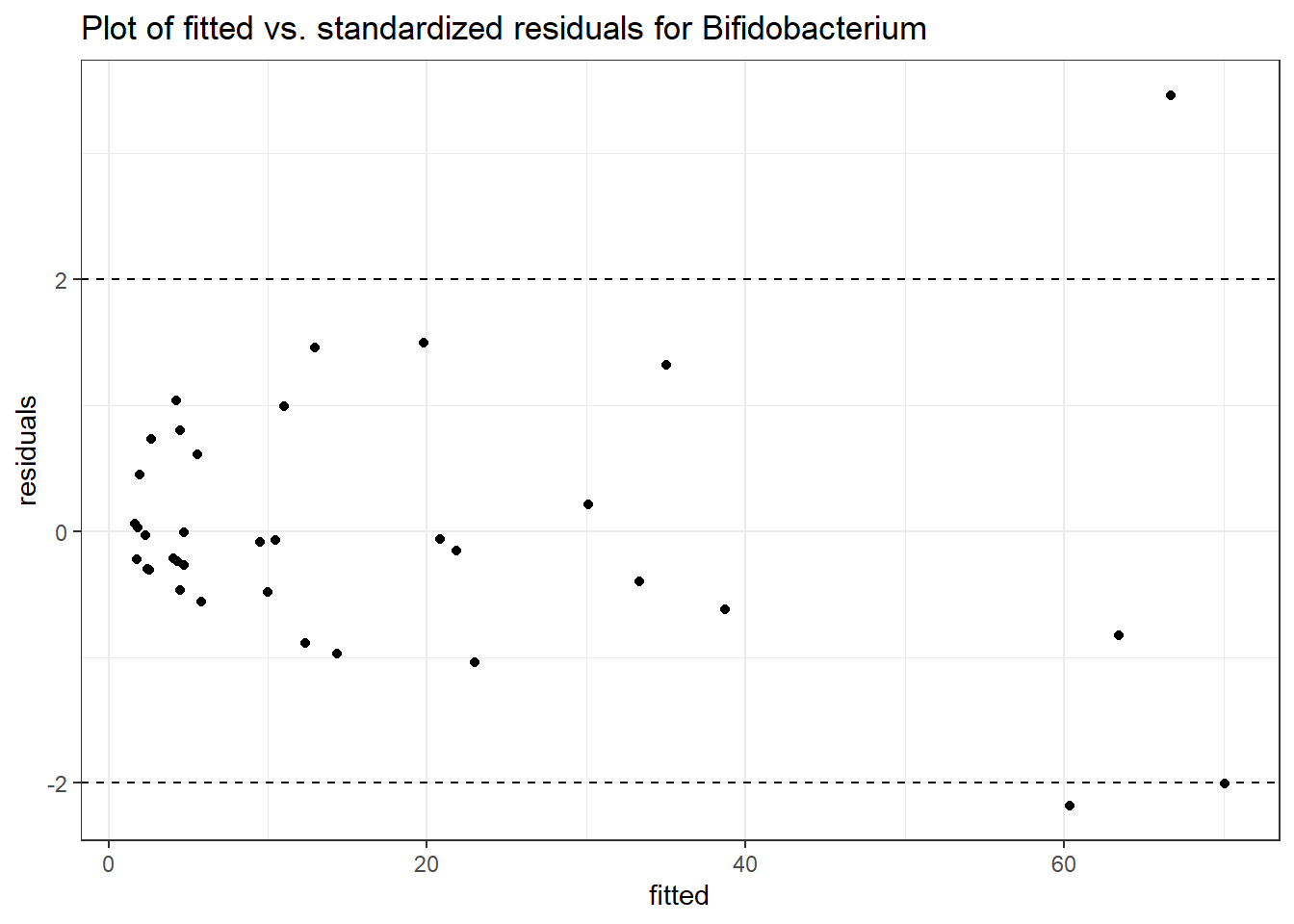
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.10788 0.58741 3.58843 0.00033
time Bifidobacterium 0.05002 0.03554 1.40735 0.15932
hispanic Bifidobacterium 0.17485 0.73203 0.23886 0.81121
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.1456
Significance test not available for random effects
#########################################
#########################################
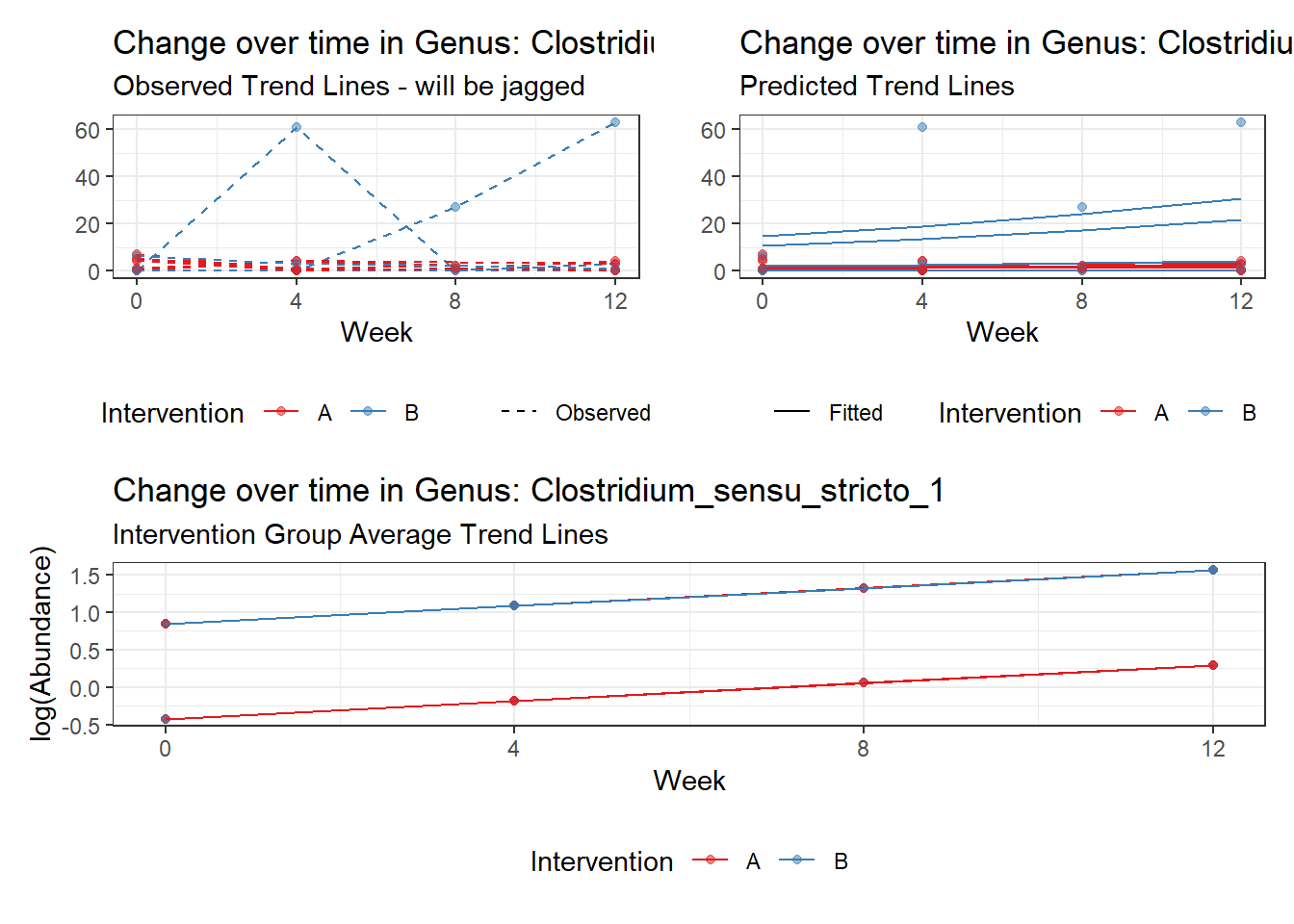
Model: Clostridium_sensu_stricto_1 ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.5760329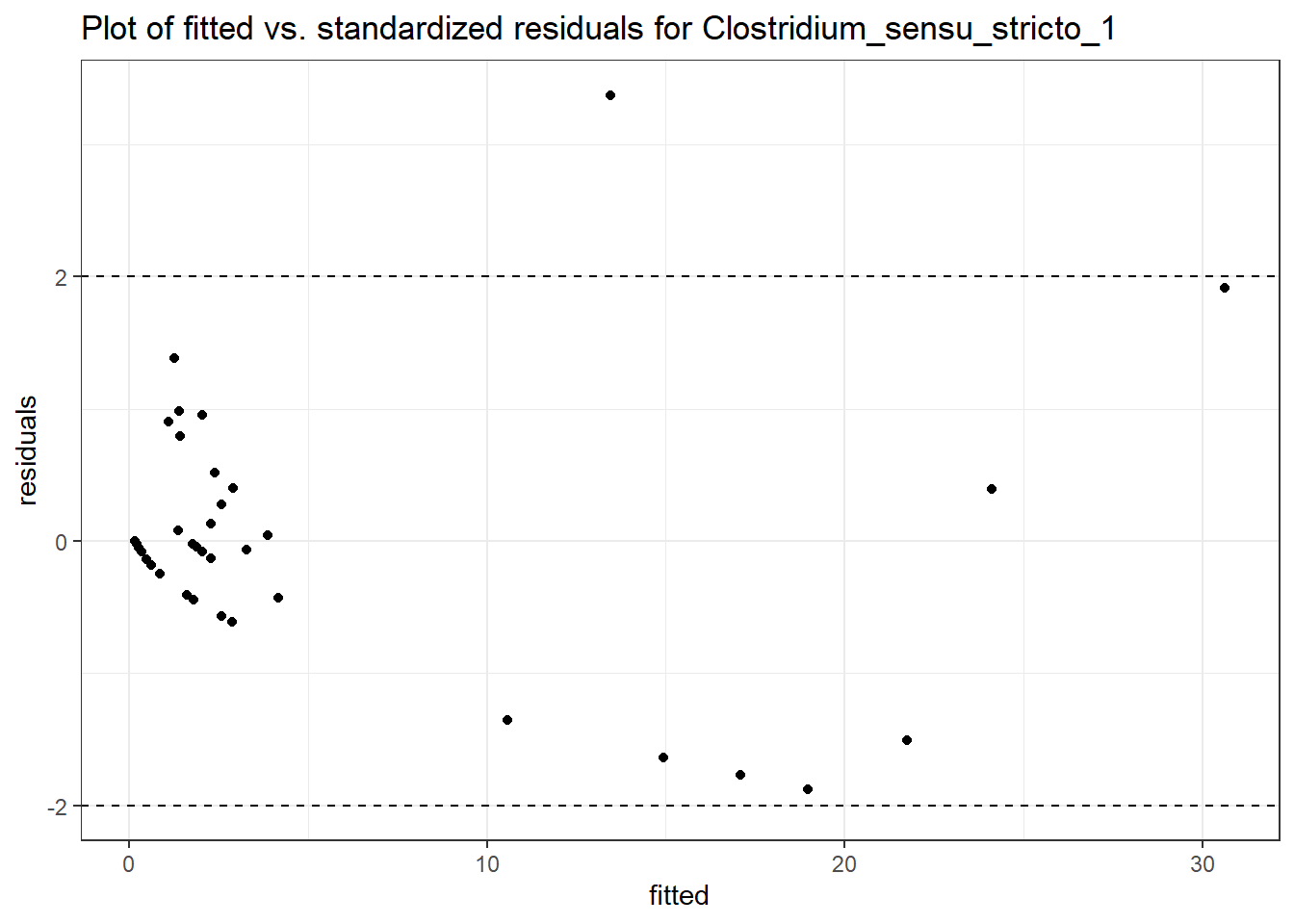
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 -0.42103 0.65987 -0.63805 0.52344
time Clostridium_sensu_stricto_1 0.23969 0.06316 3.79485 0.00015
hispanic Clostridium_sensu_stricto_1 1.26680 0.79960 1.58429 0.11313
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.1656
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.1540134
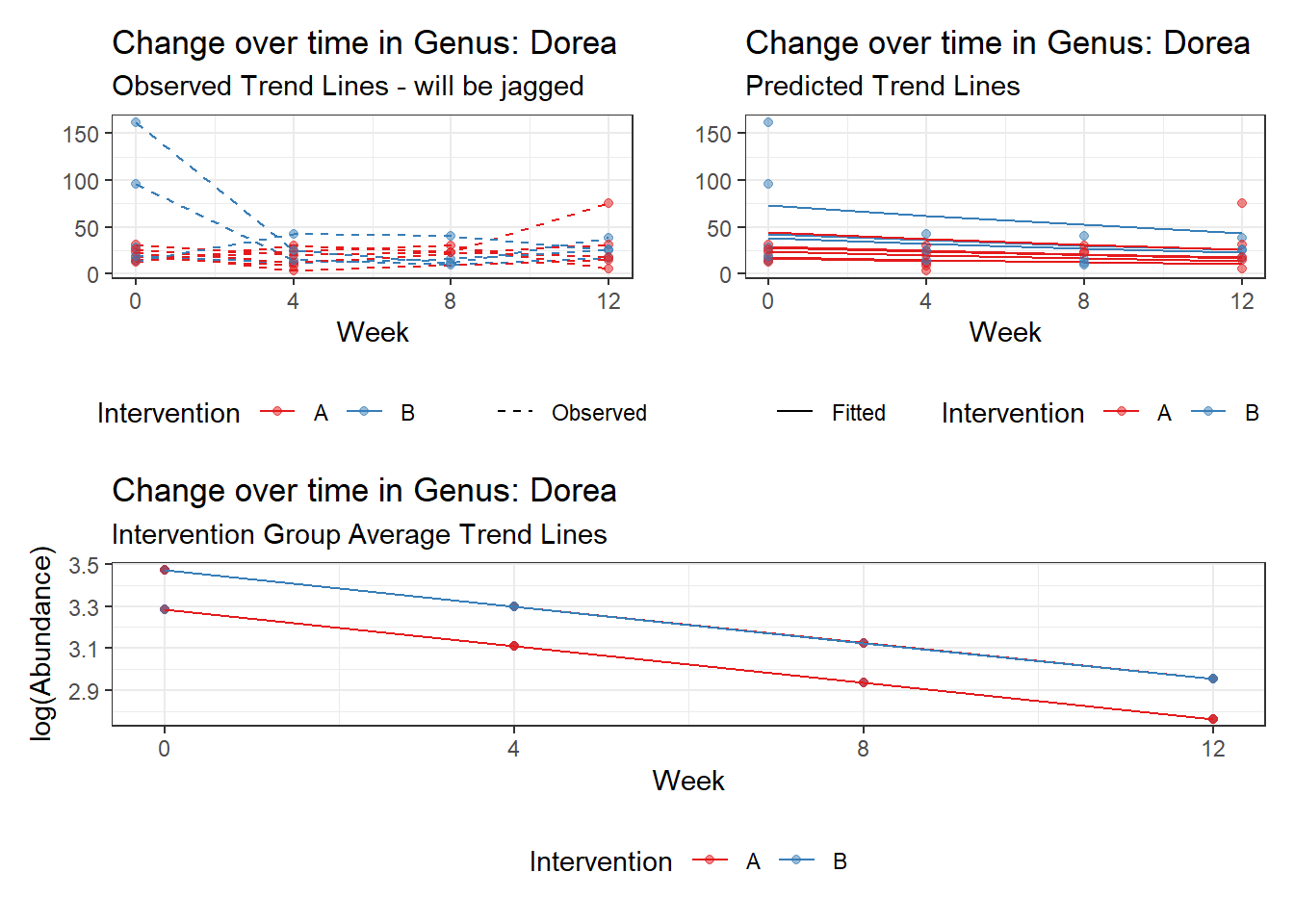
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.28367 0.22397 14.66140 0.00000
time Dorea -0.17308 0.02860 -6.05115 0.00000
hispanic Dorea 0.18859 0.27800 0.67838 0.49753
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.42668
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.2766865
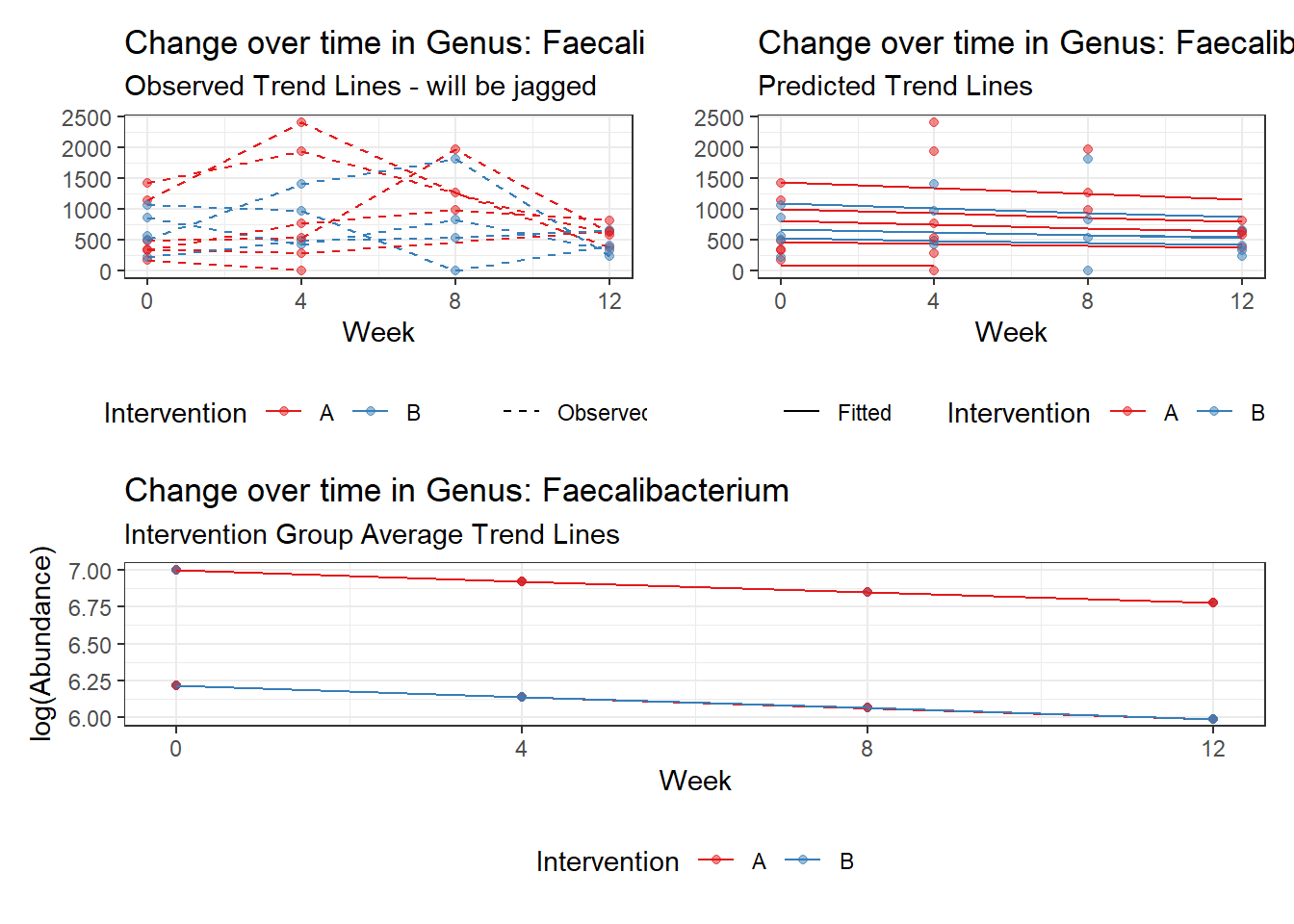
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.99992 0.30943 22.62167 0.00000
time Faecalibacterium -0.07487 0.00529 -14.16220 0.00000
hispanic Faecalibacterium -0.78455 0.38800 -2.02204 0.04317
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.61849
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.2871898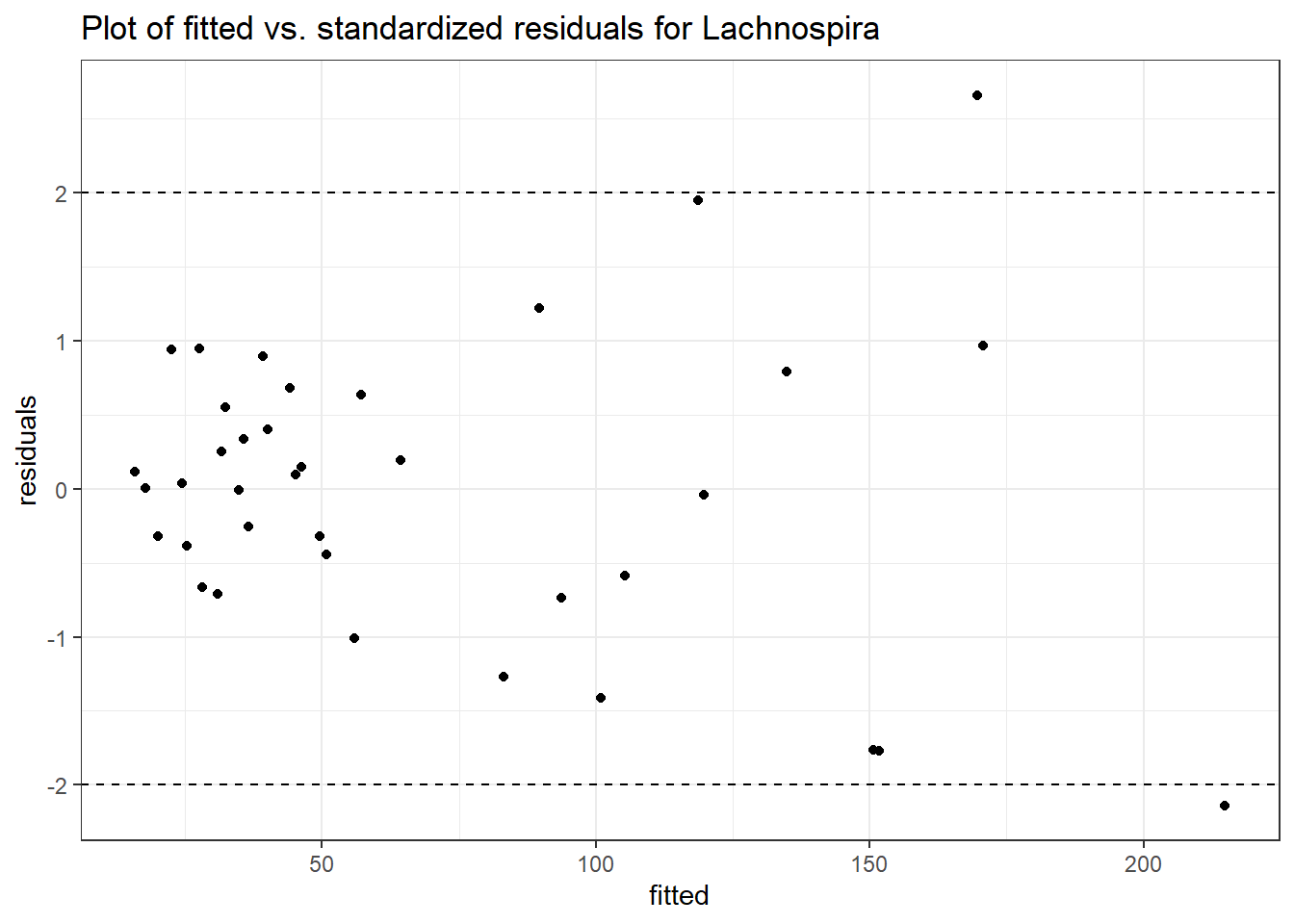

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.31759 0.31996 13.49420 0.00000
time Lachnospira 0.11818 0.01757 6.72498 0.00000
hispanic Lachnospira -0.81441 0.40158 -2.02802 0.04256
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.63474
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.8530614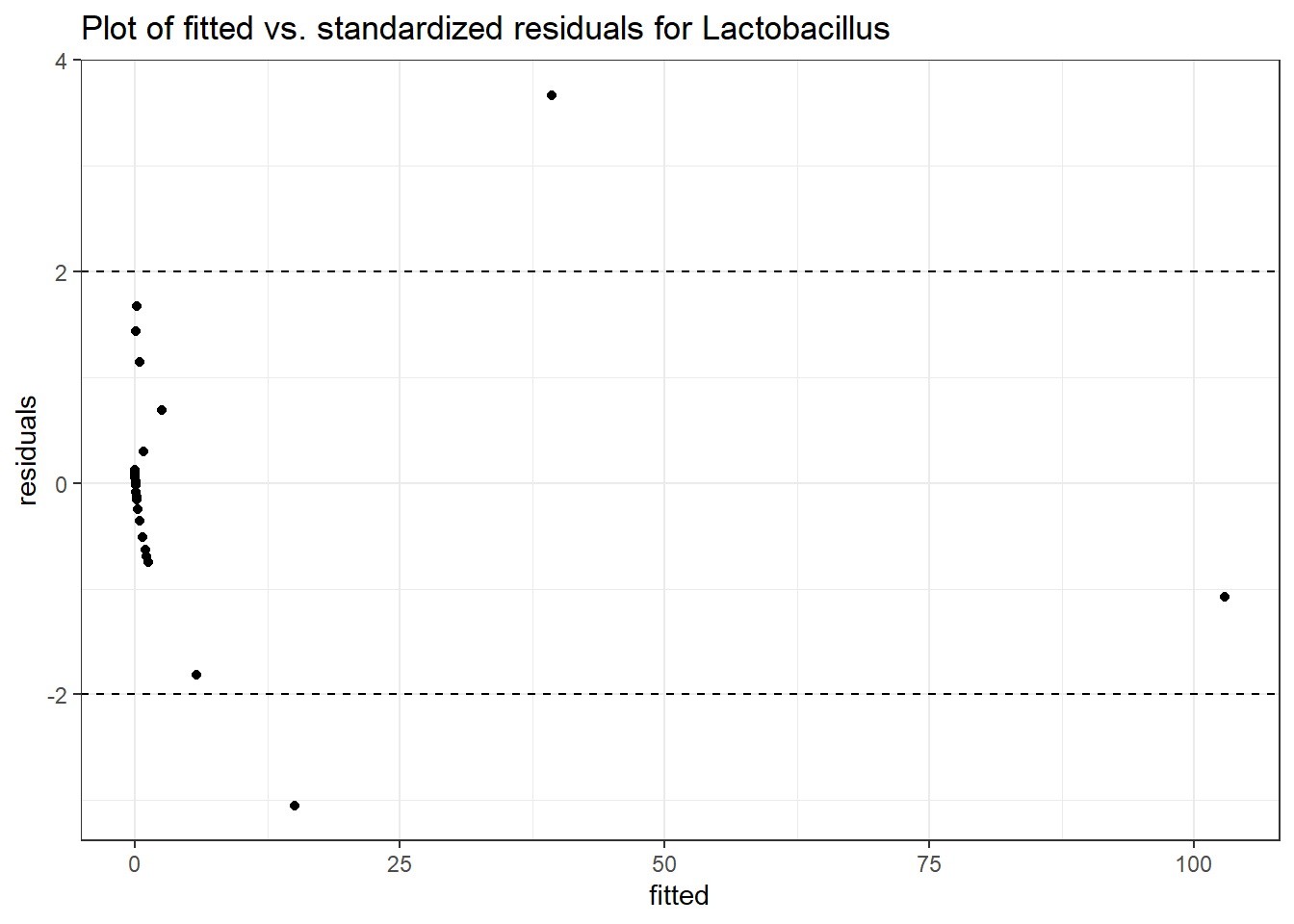
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus -0.26276 1.53162 -0.17156 0.86379
time Lactobacillus -0.96034 0.09524 -10.08340 0.00000
hispanic Lactobacillus -1.00264 1.78010 -0.56325 0.57326
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4095
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.7565355

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.29052 1.05744 -1.22042 0.22231
time Prevotella 0.10358 0.14718 0.70373 0.48160
hispanic Prevotella 0.48391 1.27445 0.37970 0.70417
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7628
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.7540662
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 1.92288 0.91177 2.10896 0.03495
time Roseburia 0.09570 0.03045 3.14263 0.00167
hispanic Roseburia 0.41539 1.12853 0.36808 0.71282
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.751
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000000698e220>
Link: poisson
Intraclass Correlation (ICC): 0.7619321

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 2.30535 0.90177 2.55647 0.01057
time Ruminococcus_1 0.25687 0.01599 16.06906 0.00000
hispanic Ruminococcus_1 1.28477 1.13362 1.13333 0.25707
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.789
Significance test not available for random effectsgenus.fit3b <- glmm_microbiome(mydata=microbiome_data,model.number=3,
taxa.level="Genus", taxa.subset = backNames,
link="negbinom",
model="1 + time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced
Intraclass Correlation (ICC): 0.8519852

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 3.82266 1.16237 3.28867 0.00101
time Akkermansia 0.00195 0.00938 0.20773 0.83544
hispanic Akkermansia 1.08648 1.46487 0.74169 0.45827
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3992
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinom
Intraclass Correlation (ICC): 0.1779314
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 8.08994 0.25500 31.72470 0.00000
time Bacteroides -0.06717 0.04497 -1.49389 0.13520
hispanic Bacteroides -0.60635 0.31200 -1.94340 0.05197
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.46523
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinom
Intraclass Correlation (ICC): 0.3759523
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.41192 0.80989 2.97809 0.00290
time Bifidobacterium 0.00520 0.28759 0.01809 0.98557
hispanic Bifidobacterium -0.02503 0.77634 -0.03224 0.97428
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.77617
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 2.643847e-14
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.20528 0.58987 0.34801 0.72783
time Clostridium_sensu_stricto_1 0.13363 0.29653 0.45064 0.65225
hispanic Clostridium_sensu_stricto_1 1.67539 0.69795 2.40043 0.01638
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.626e-07
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinom
Intraclass Correlation (ICC): 0.005543064
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.26866 0.23391 13.97398 0.00000
time Dorea -0.12645 0.09909 -1.27619 0.20189
hispanic Dorea 0.34292 0.25193 1.36115 0.17347
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.074659
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 2.450093e-12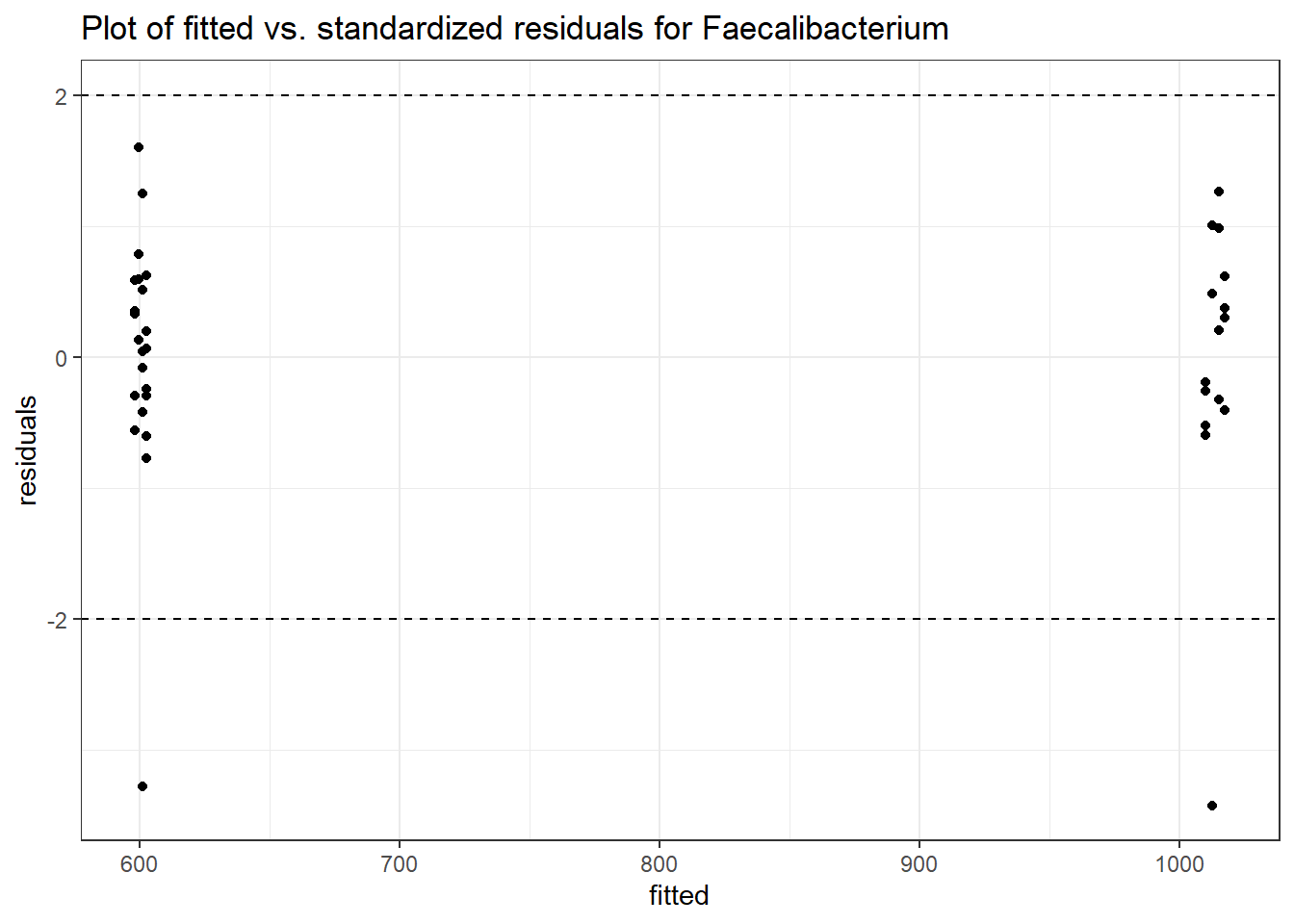
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.92530 0.30090 23.01543 0.0000
time Faecalibacterium -0.00242 0.14956 -0.01616 0.9871
hispanic Faecalibacterium -0.52422 0.31540 -1.66206 0.0965
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.5653e-06
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reached
Warning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, : NaNs
produced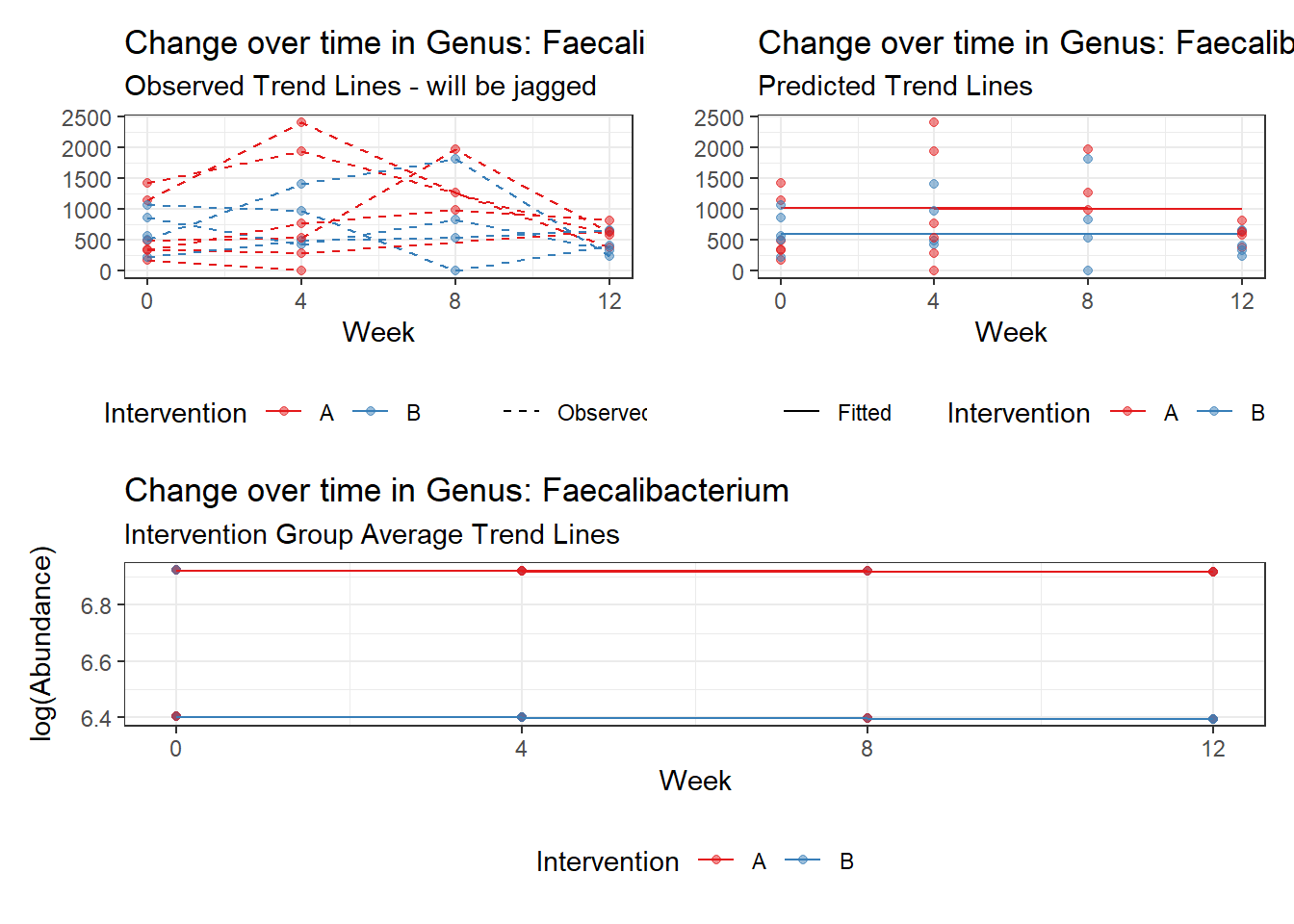
Intraclass Correlation (ICC): 0.2871283
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.31649 0.32007 13.48599 0.00000
time Lachnospira 0.11924 0.01779 6.70398 0.00000
hispanic Lachnospira -0.81481 0.40179 -2.02795 0.04257
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.63465
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00774533 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 4.020719e-10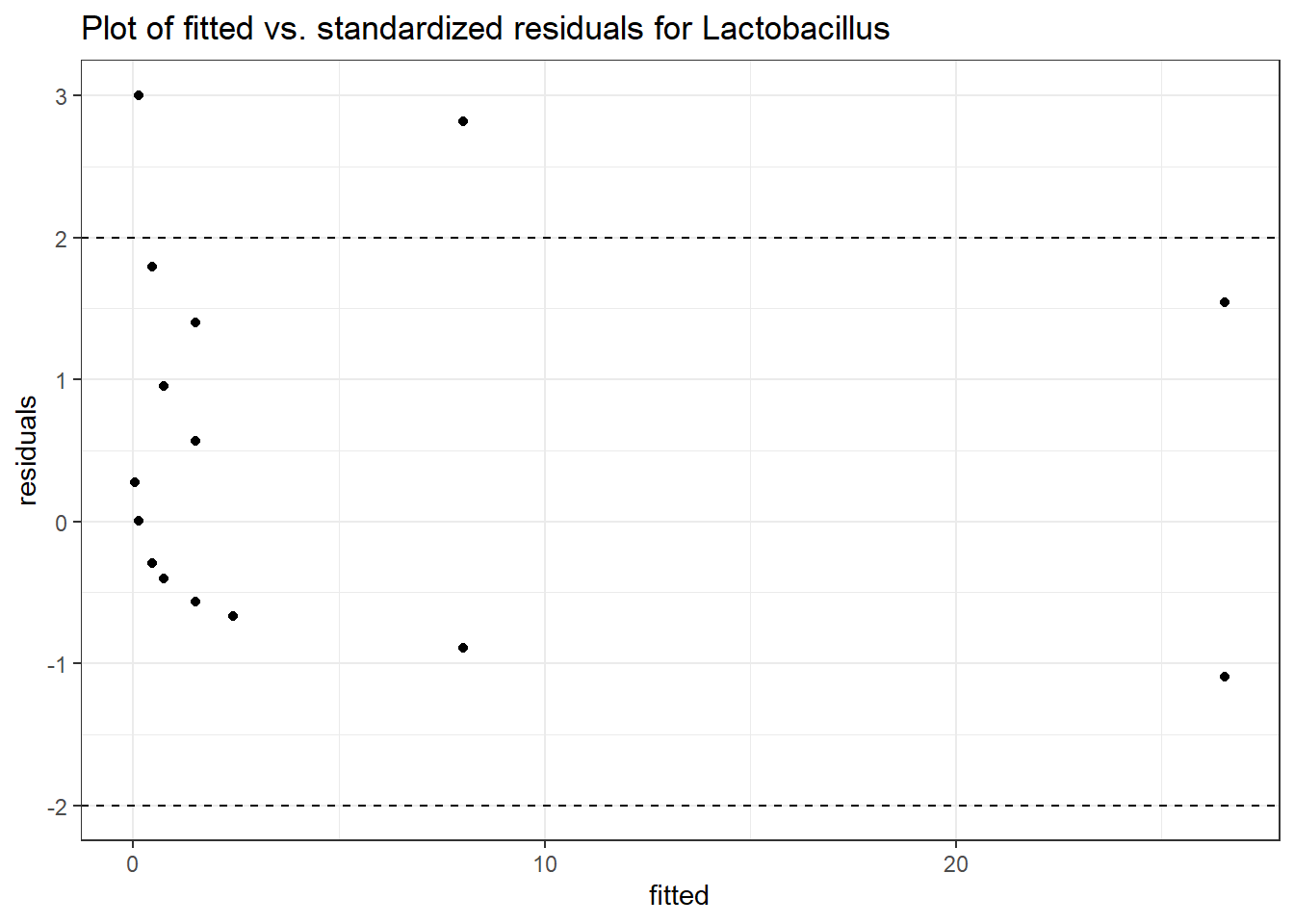
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus 3.27886 1.18473 2.76759 0.00565
time Lactobacillus -1.19822 0.75702 -1.58280 0.11347
hispanic Lactobacillus -2.86205 1.31647 -2.17404 0.02970
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.0052e-05
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 5.634536e-13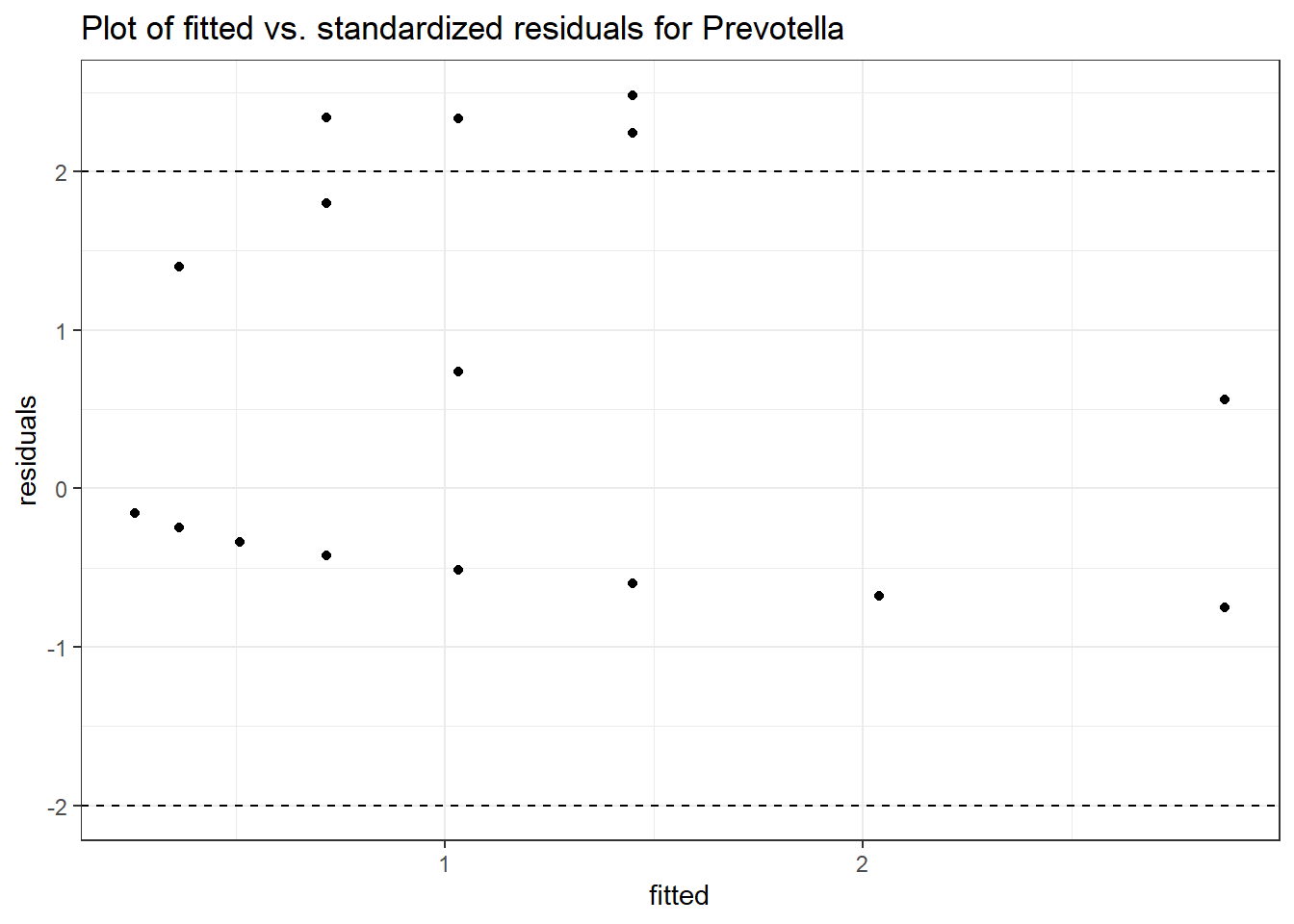
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.35712 1.76106 -0.77063 0.44093
time Prevotella 0.34101 0.64510 0.52861 0.59708
hispanic Prevotella 1.38721 1.51495 0.91568 0.35983
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 7.5064e-07
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0135518 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular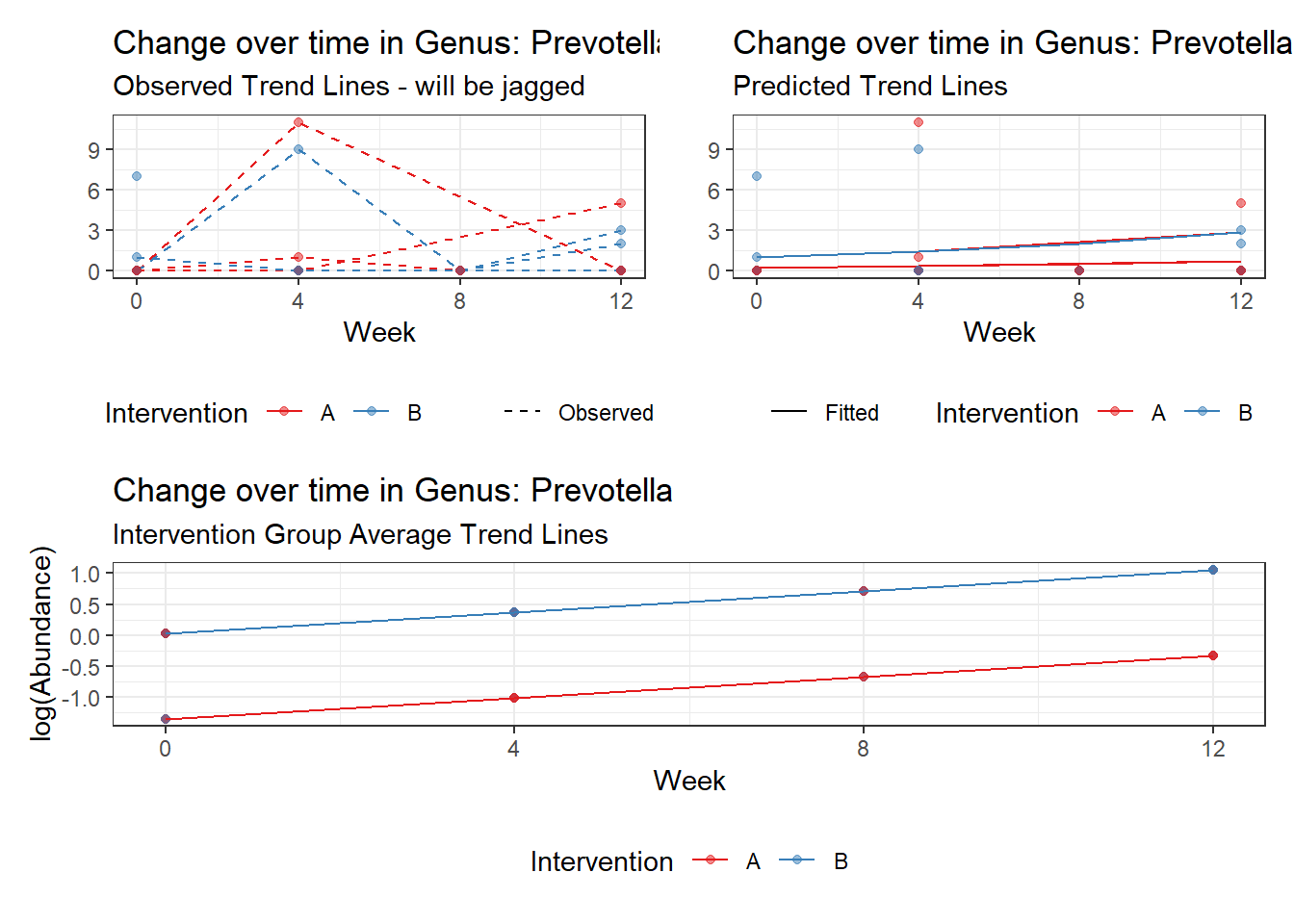
Intraclass Correlation (ICC): 0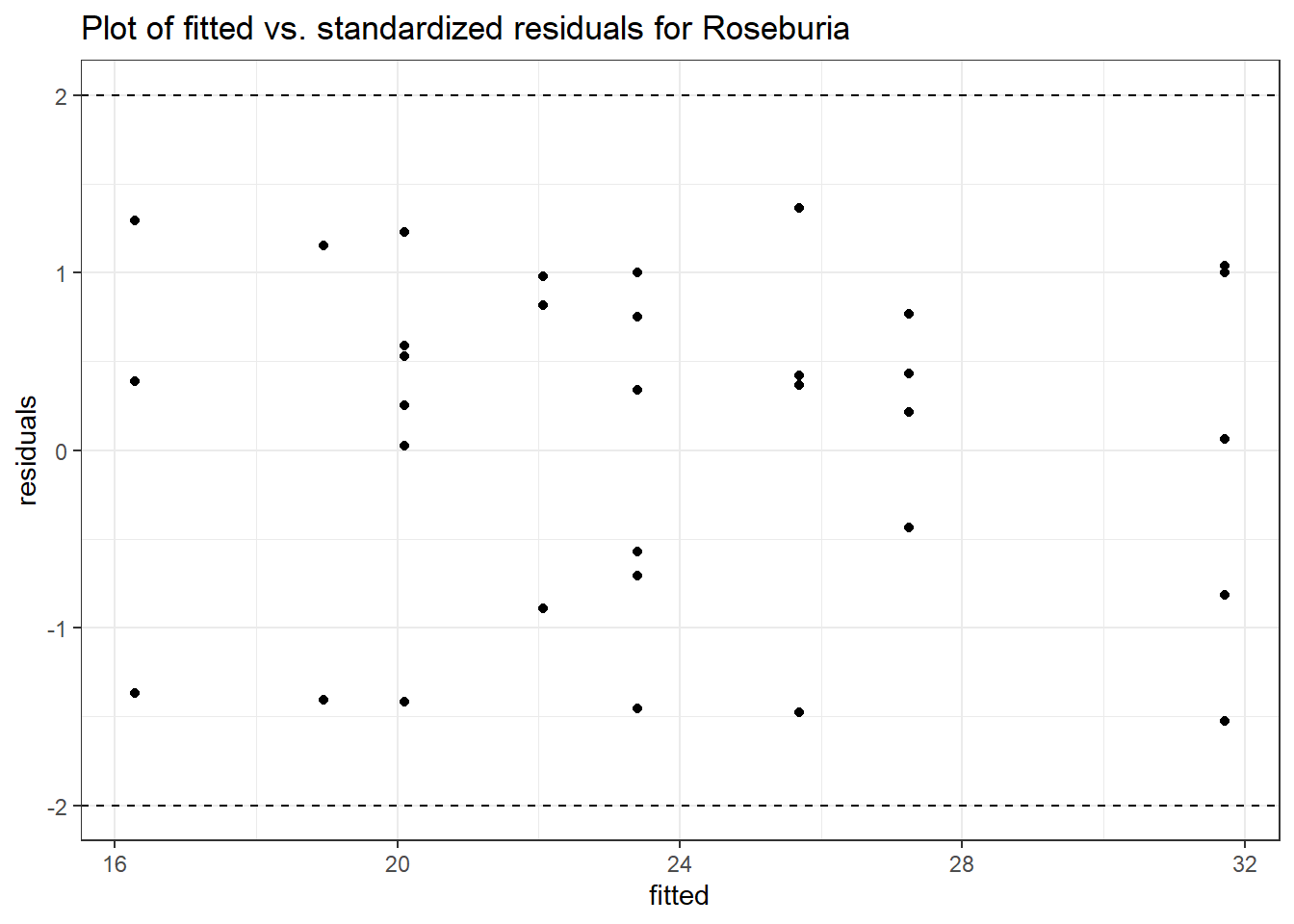
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 2.79014 0.51908 5.37513 0.00000
time Roseburia 0.15202 0.20824 0.73003 0.46537
hispanic Roseburia 0.21075 0.51174 0.41183 0.68046
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x0000000047f50298>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00777325 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 2.166005e-10

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 3.36130 0.51709 6.50043 0.00000
time Ruminococcus_1 0.27986 0.22569 1.24003 0.21496
hispanic Ruminococcus_1 0.95368 0.54067 1.76388 0.07775
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4717e-05
Significance test not available for random effectsControlling for the effect of time, Ethnicity was not significant for all Genus.
Effect of Intervention
Next, we invested the effect of intervention, however, the results are parsed between which covariates are included in the model.
Intervention was coded as (group A = 0, and group B = 1). So, the results are interpretted as the effect of being in intervention group B.
Covariate(s): Time
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Intervention)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
The effect of intervention is the main outcome of this study.
genus.fit4 <- glmm_microbiome(mydata=microbiome_data,model.number=4,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + intB + time + (1|SubjectID)")
#########################################
#########################################
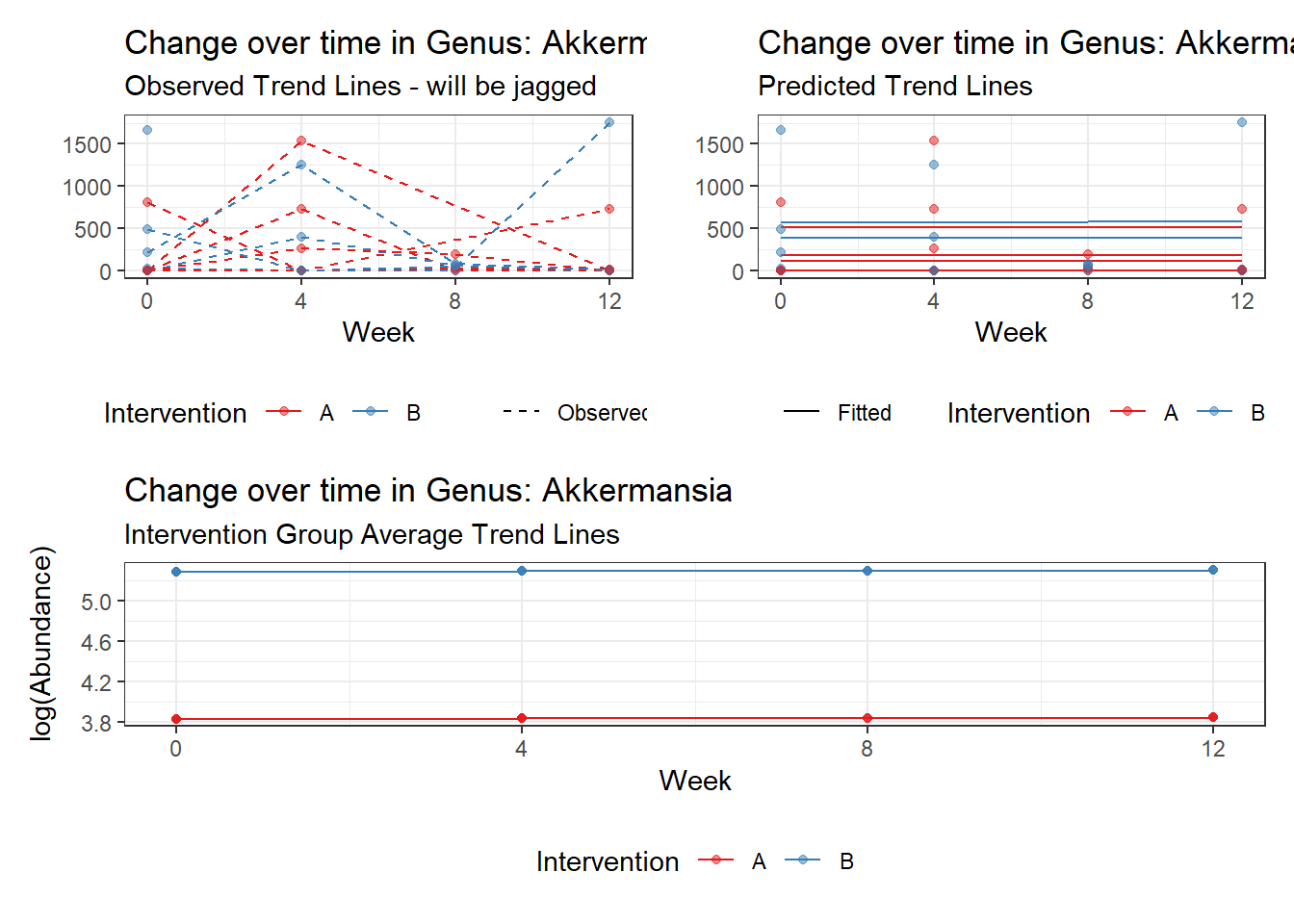
Model: Akkermansia ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.8508851
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 3.83630 0.99216 3.86662 0.00011
intB Akkermansia 1.44885 1.45824 0.99356 0.32044
time Akkermansia 0.00500 0.00920 0.54381 0.58657
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3888
Significance test not available for random effects
#########################################
#########################################
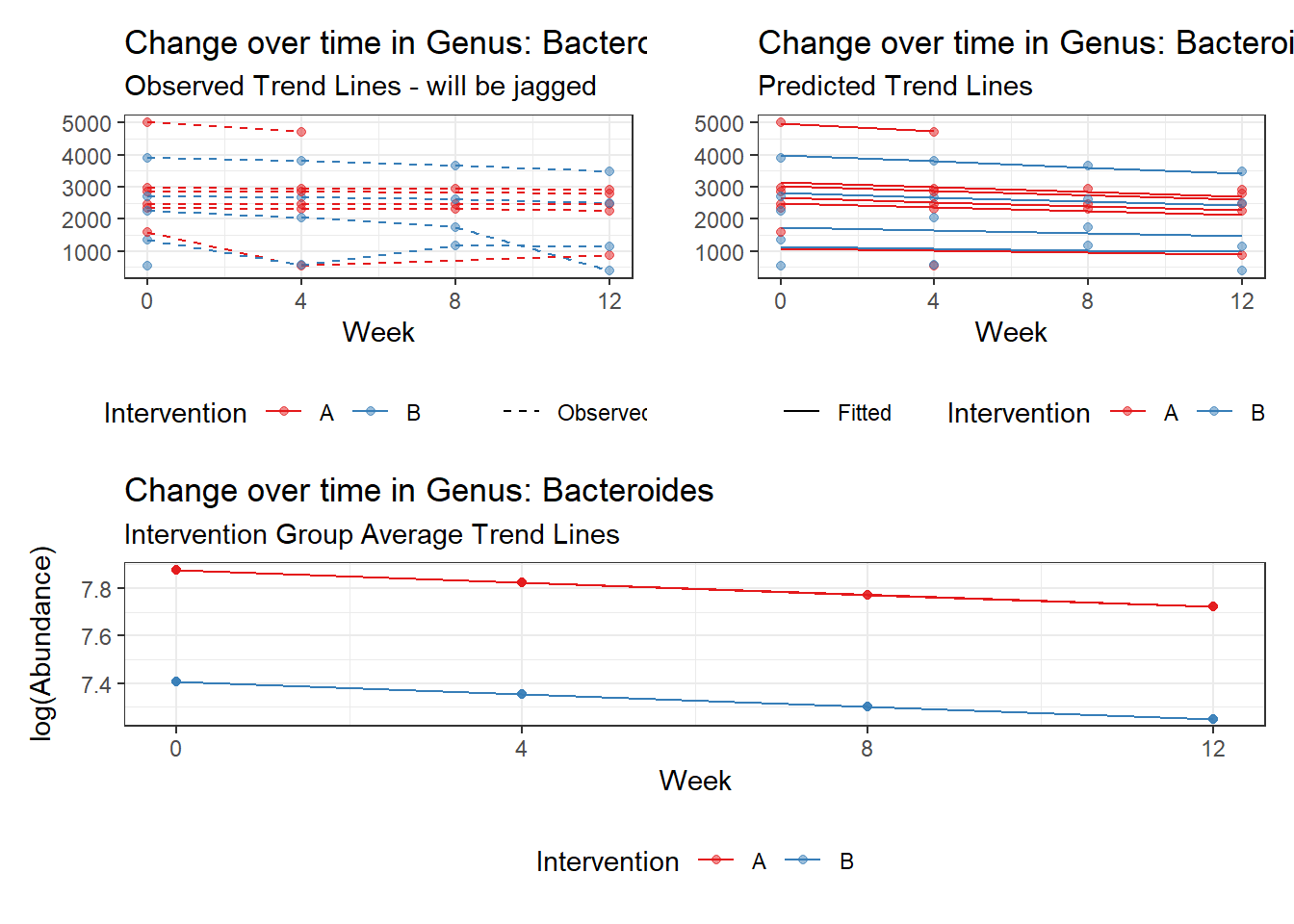
Model: Bacteroides ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.2529351
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.87622 0.23763 33.14512 0.00000
intB Bacteroides -0.47061 0.35251 -1.33501 0.18187
time Bacteroides -0.05160 0.00312 -16.55687 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.58187
Significance test not available for random effects
#########################################
#########################################
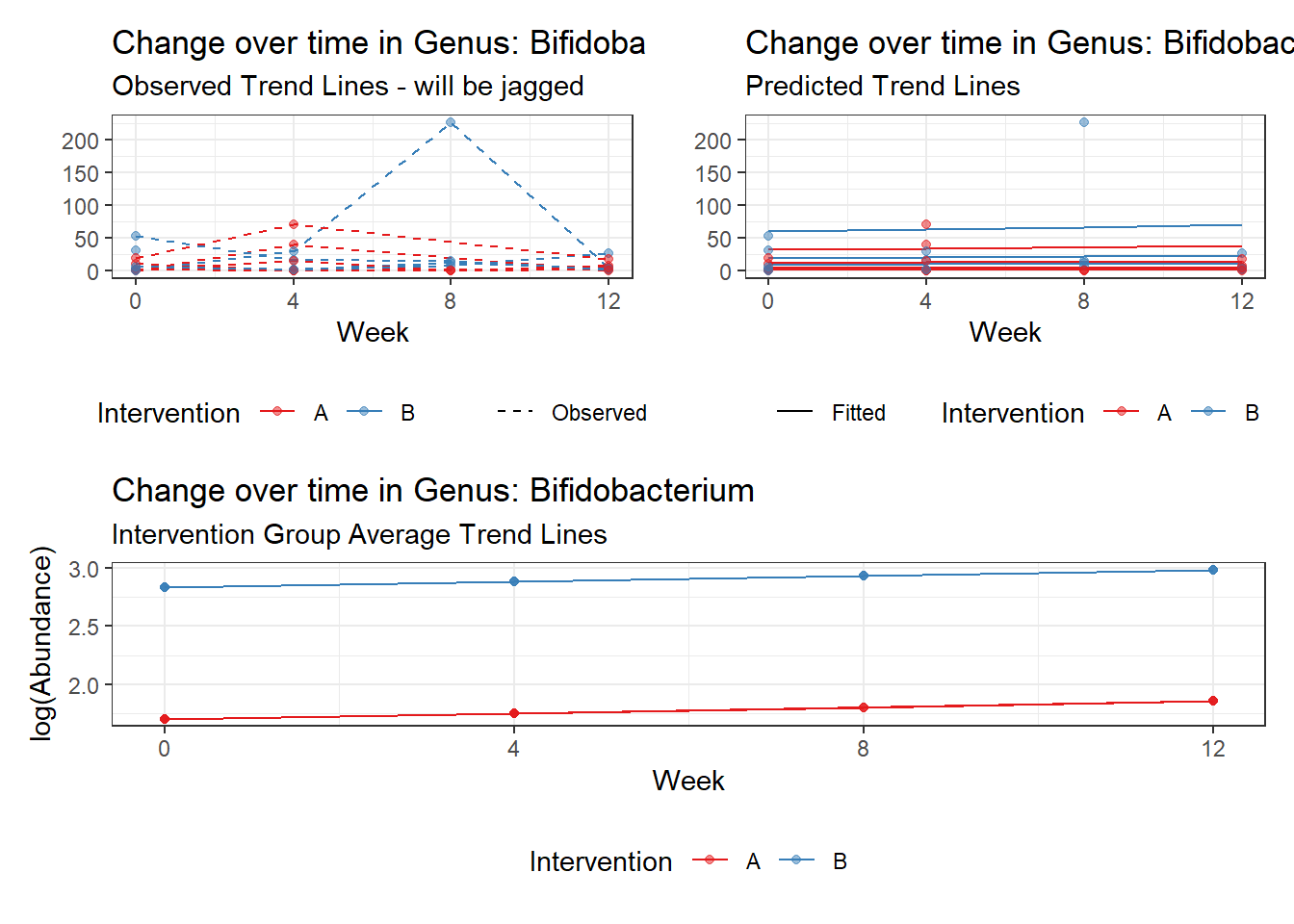
Model: Bifidobacterium ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.5025902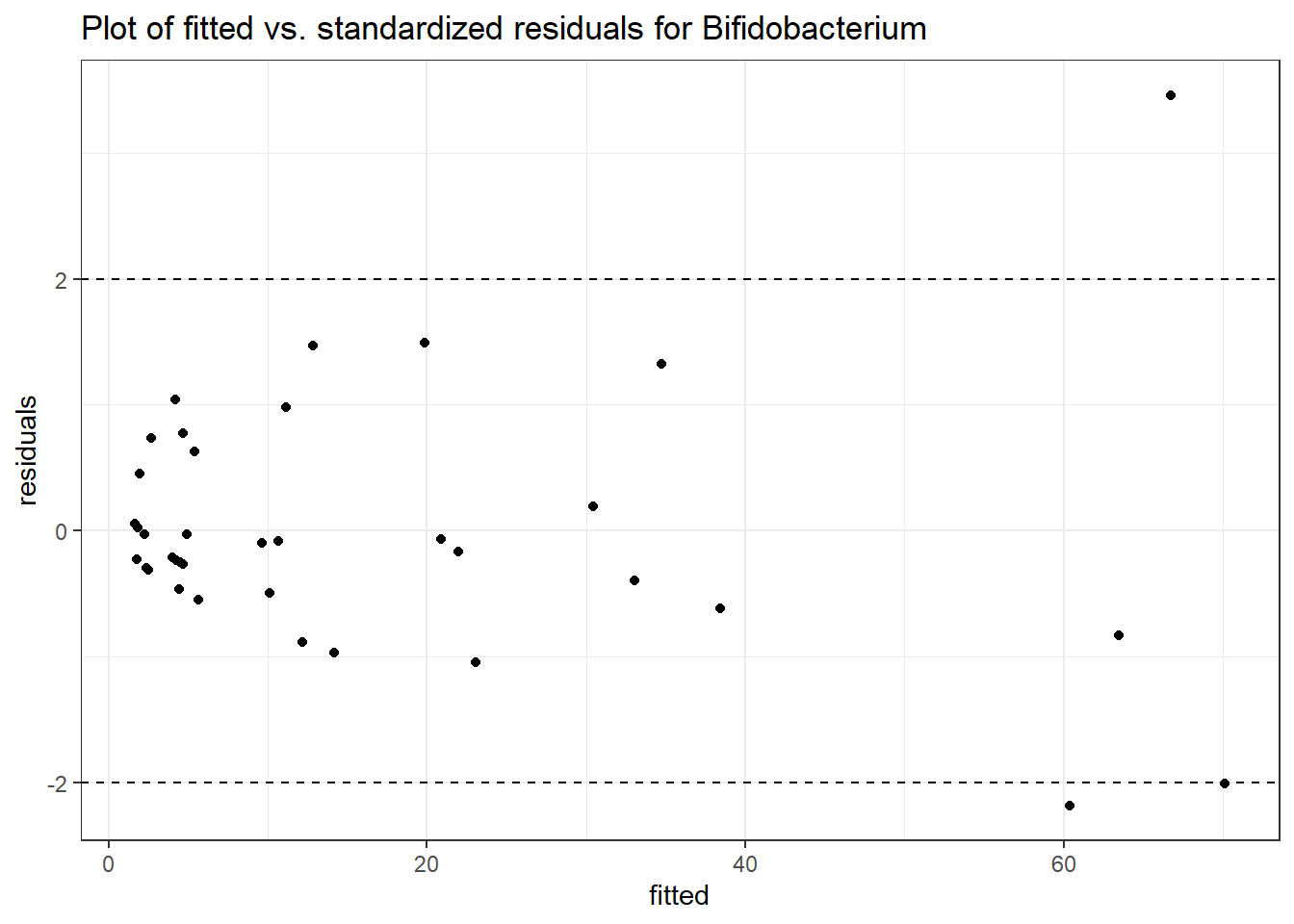
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 1.70526 0.42717 3.99203 0.00007
intB Bifidobacterium 1.12673 0.62217 1.81095 0.07015
time Bifidobacterium 0.04980 0.03554 1.40128 0.16113
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0052
Significance test not available for random effects
#########################################
#########################################
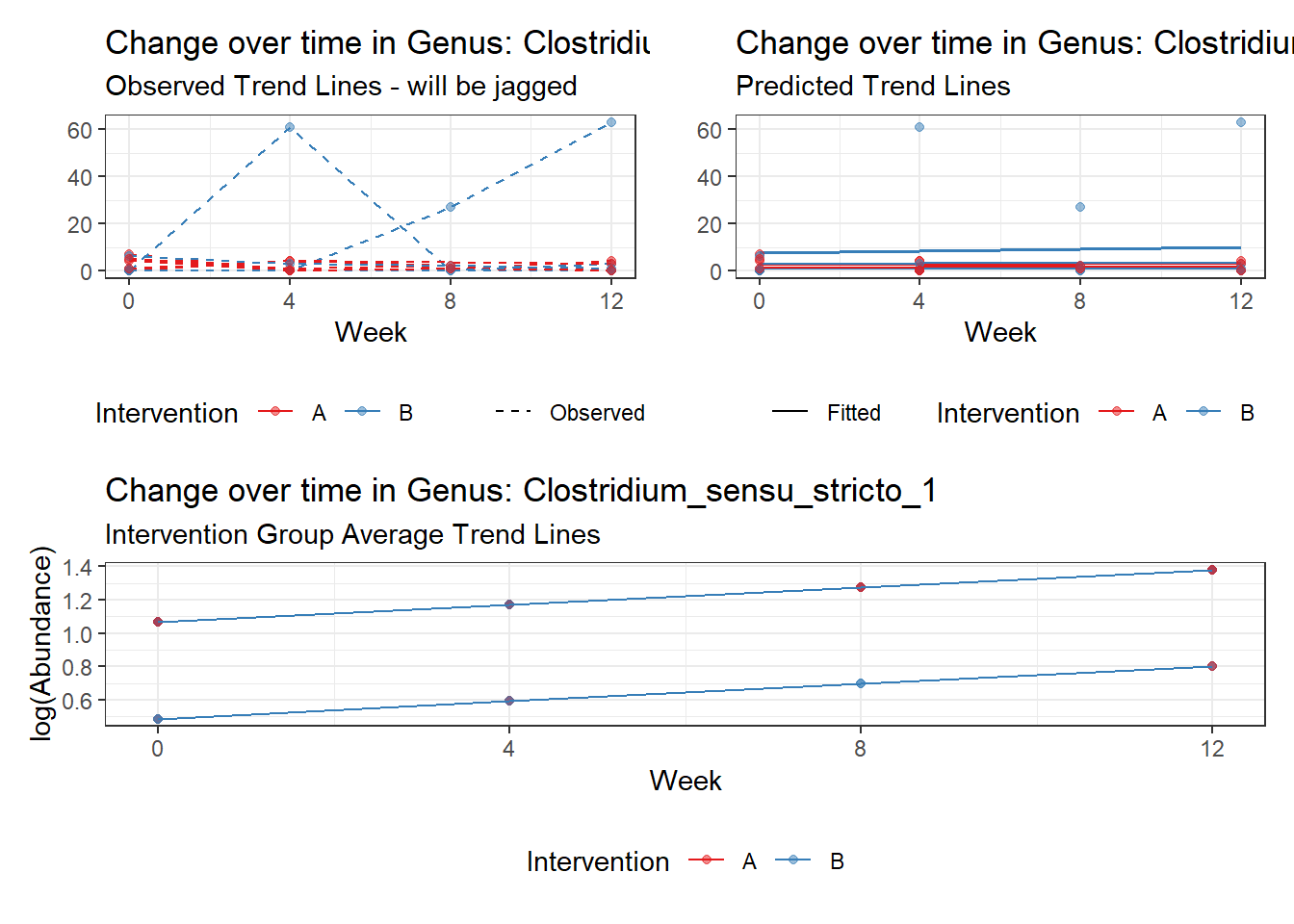
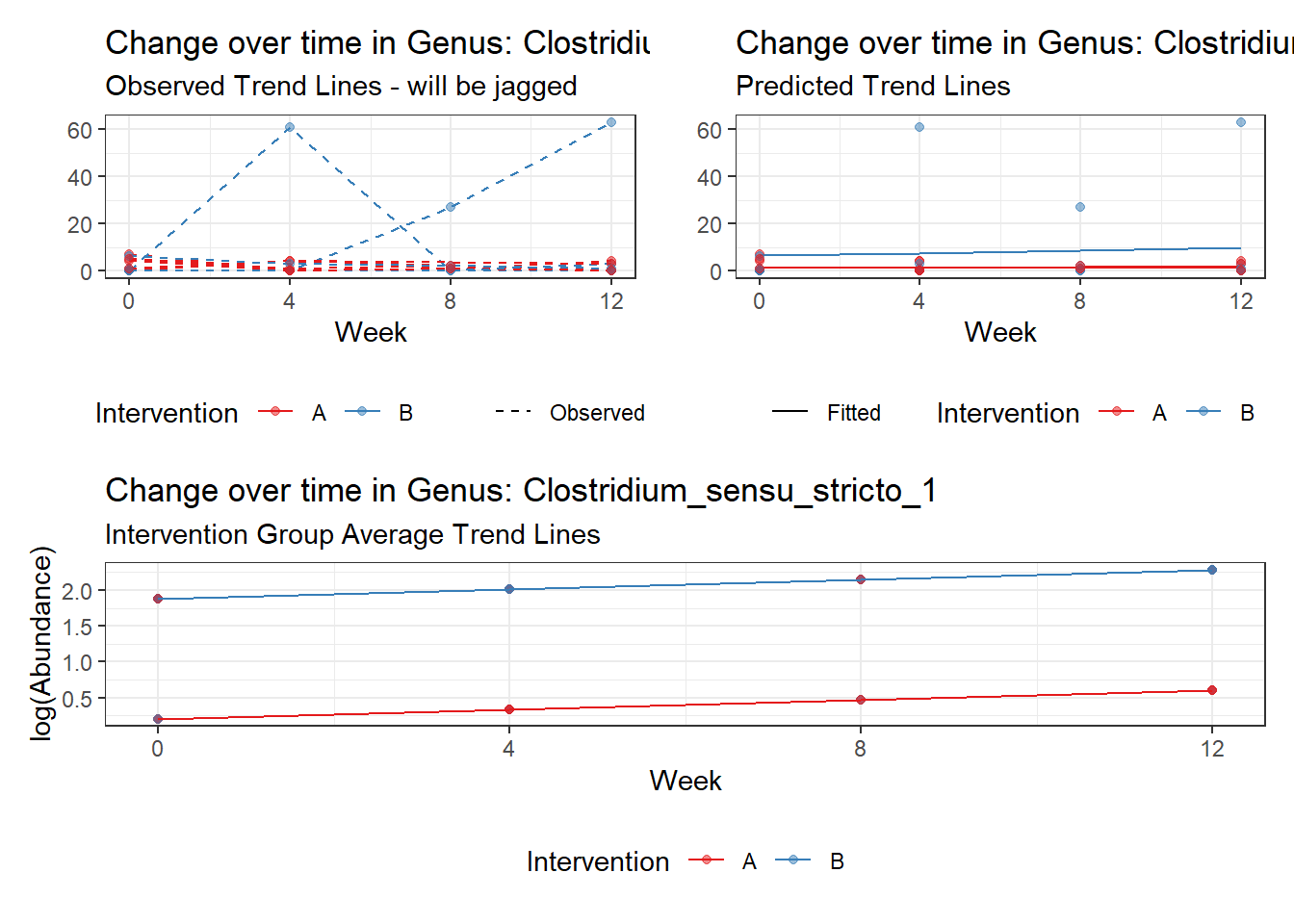
Model: Clostridium_sensu_stricto_1 ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.5985013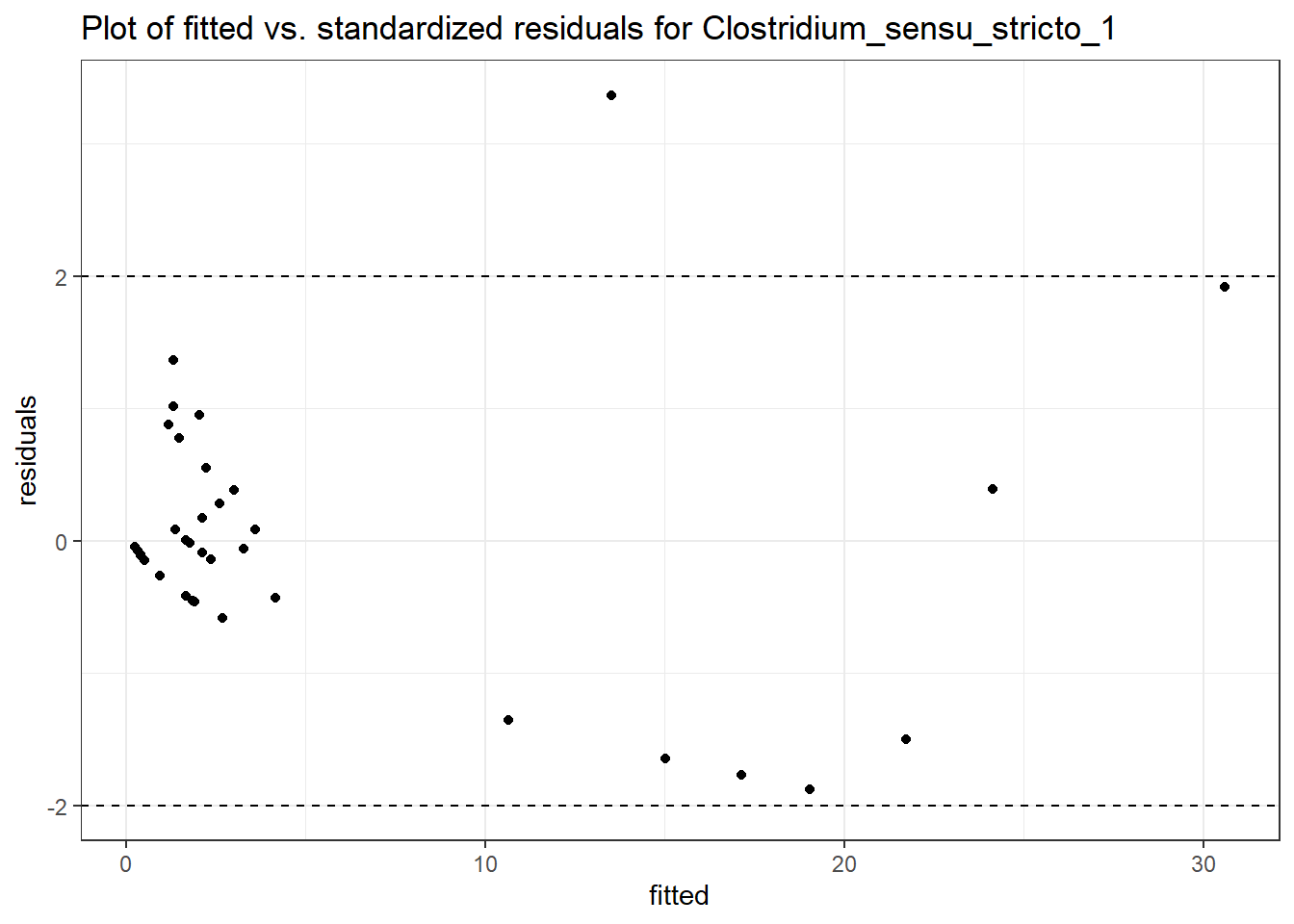

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 -0.02813 0.55311 -0.05085 0.95945
intB Clostridium_sensu_stricto_1 0.90230 0.80074 1.12683 0.25982
time Clostridium_sensu_stricto_1 0.23751 0.06305 3.76735 0.00016
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.2209
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.1234956

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.19880 0.16526 19.35643 0.00000
intB Dorea 0.45197 0.23878 1.89285 0.05838
time Dorea -0.17253 0.02859 -6.03508 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.37536
Significance test not available for random effects
#########################################
#########################################
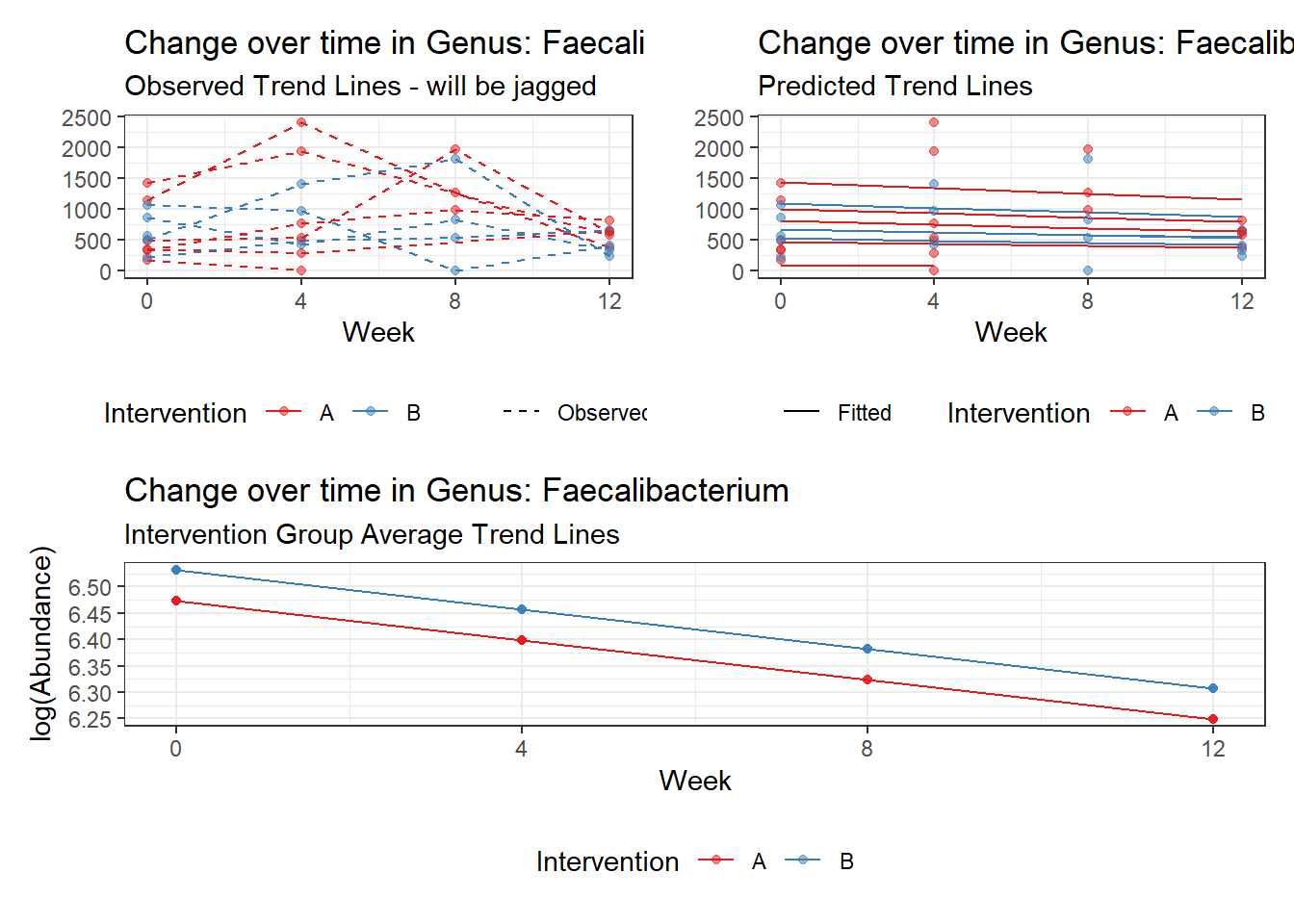
Model: Faecalibacterium ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.344455
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.47432 0.29634 21.84738 0.00000
intB Faecalibacterium 0.05749 0.43932 0.13085 0.89589
time Faecalibacterium -0.07486 0.00529 -14.16058 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.72488
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
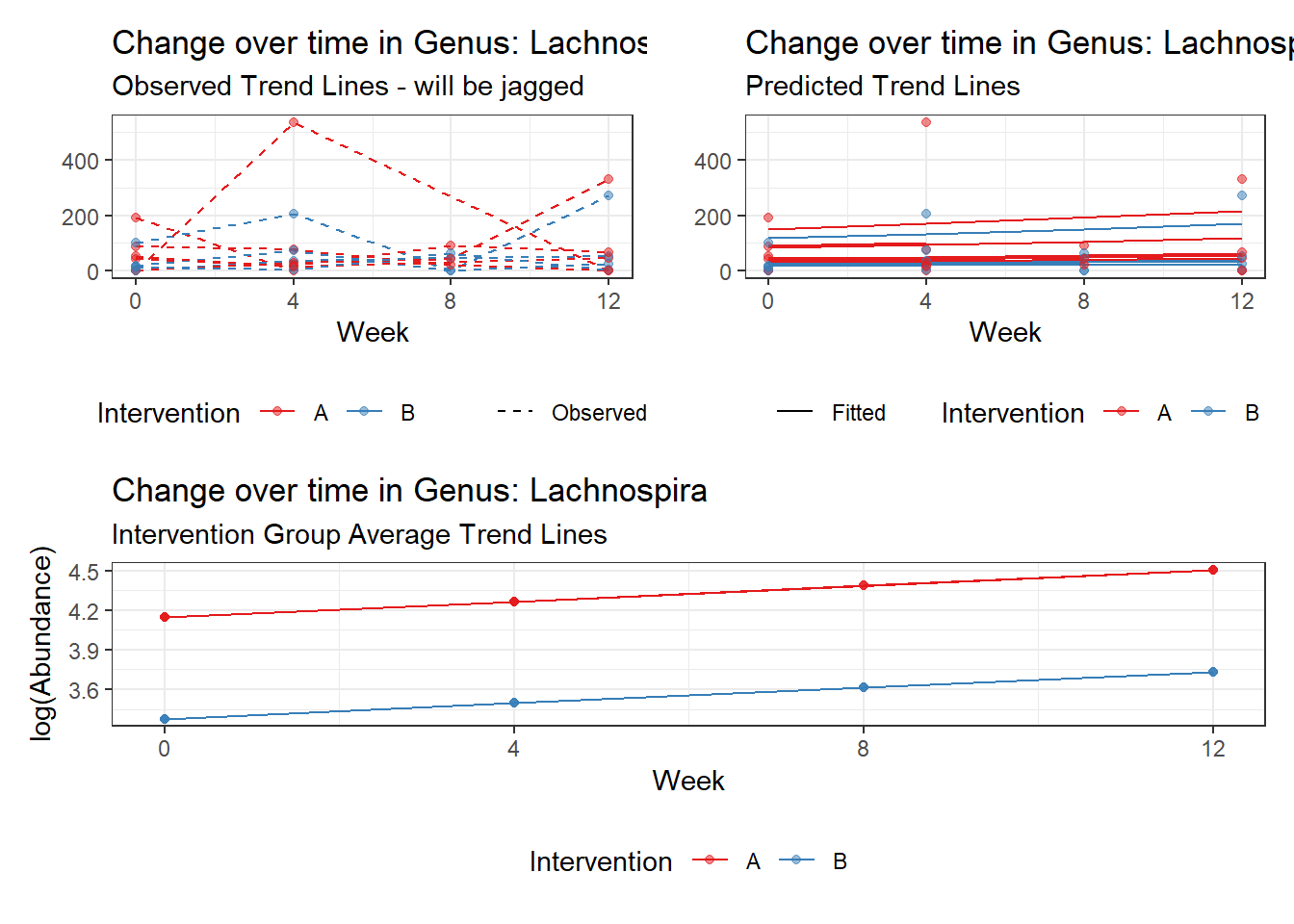
Intraclass Correlation (ICC): 0.2927057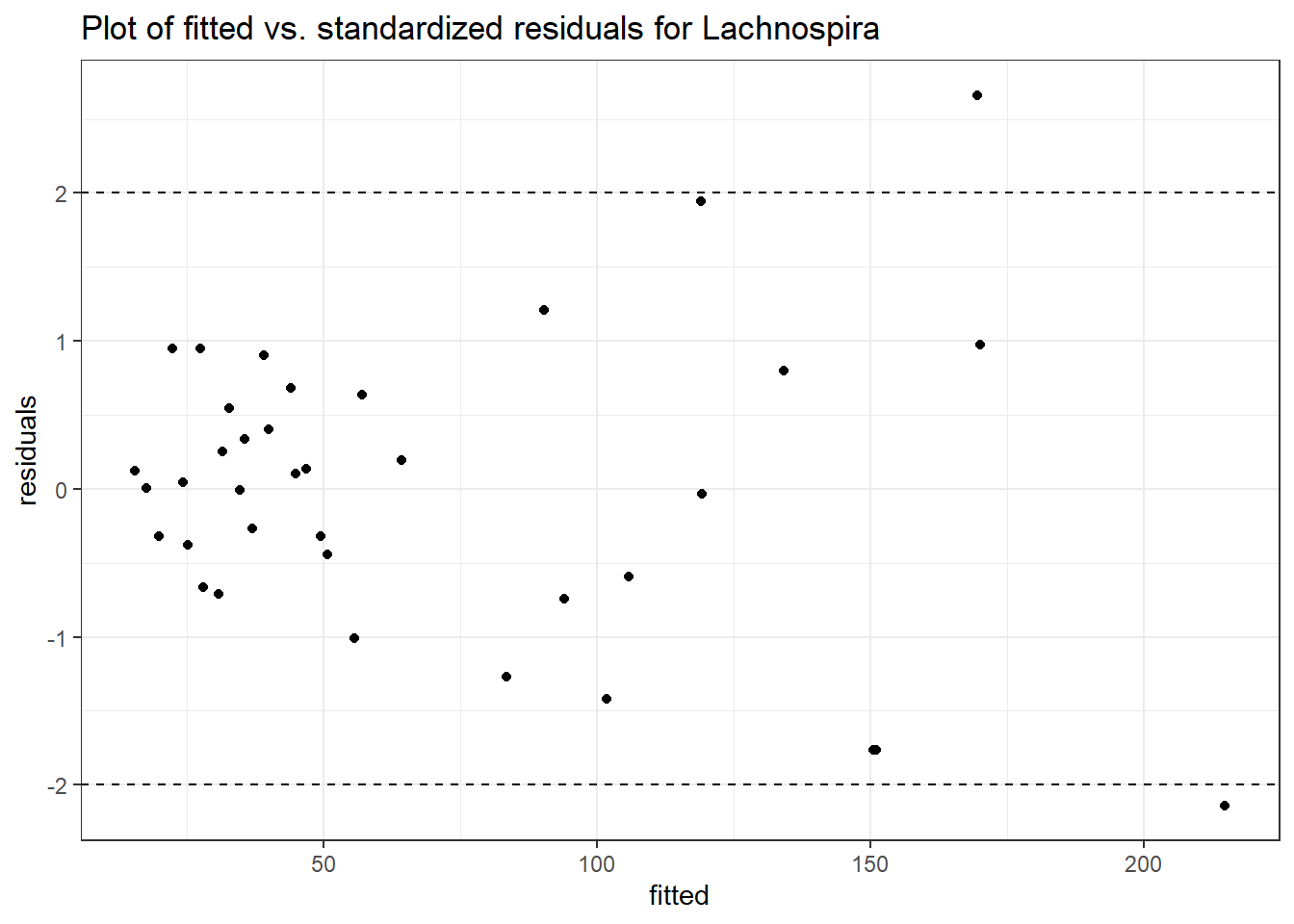
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.14833 0.26528 15.63754 0.00000
intB Lachnospira -0.77161 0.39510 -1.95293 0.05083
time Lachnospira 0.11863 0.01758 6.74963 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.6433
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
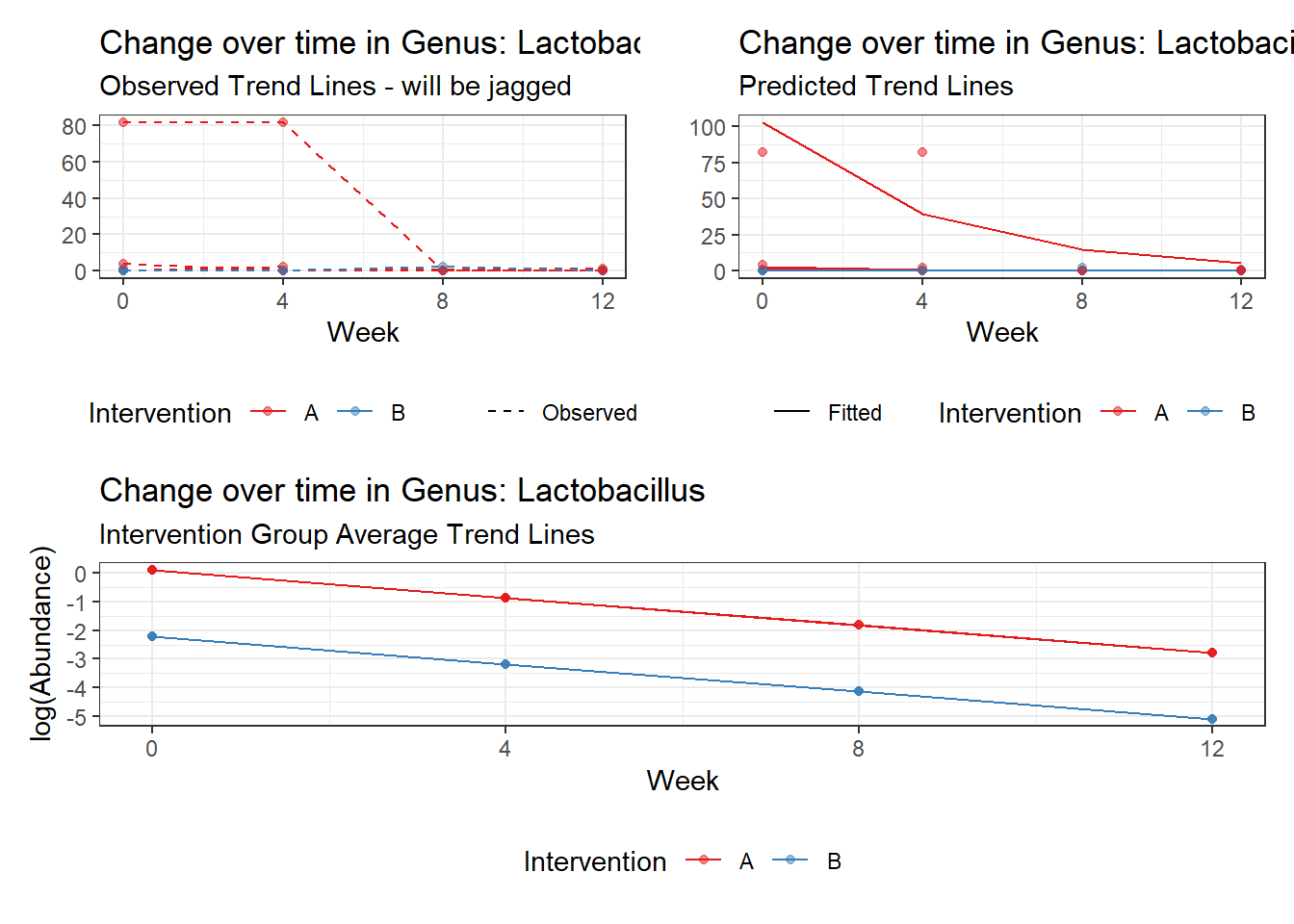
Intraclass Correlation (ICC): 0.8357546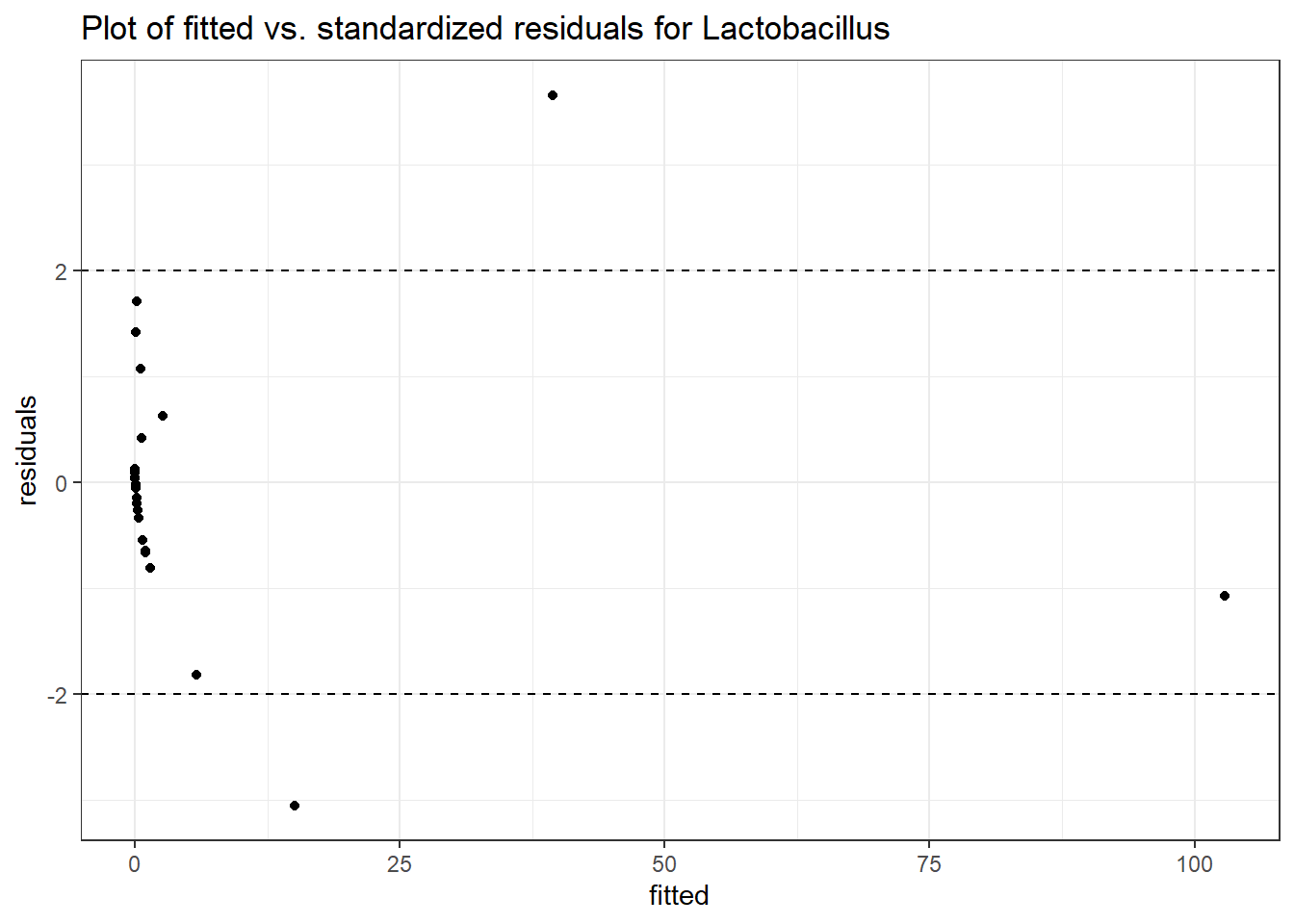
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus 0.11350 1.08713 0.10441 0.91685
intB Lactobacillus -2.32857 1.67987 -1.38616 0.16570
time Lactobacillus -0.96020 0.09525 -10.08089 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.2558
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.757277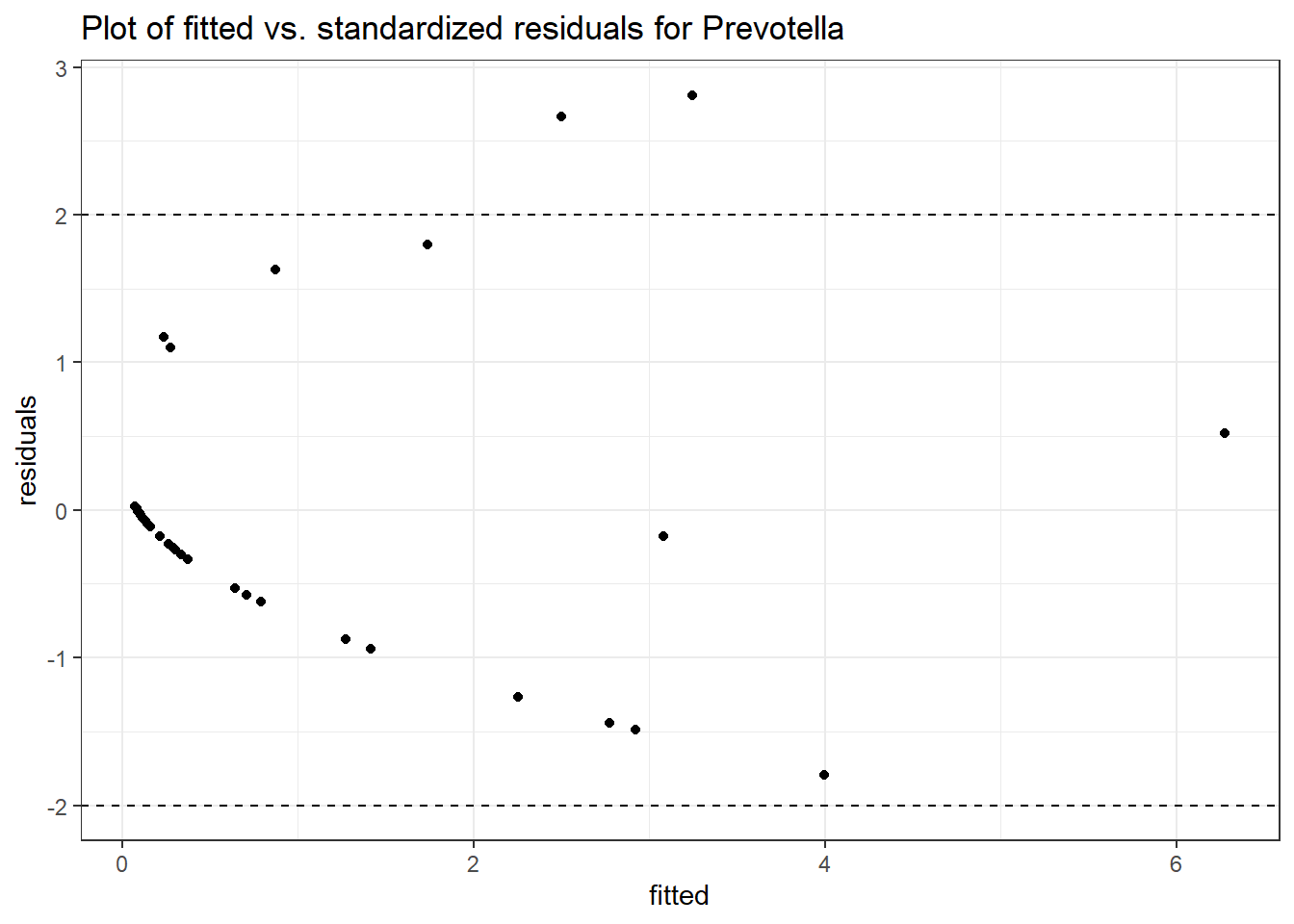
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.55221 1.00217 -1.54885 0.12142
intB Prevotella 1.13051 1.25230 0.90275 0.36666
time Prevotella 0.10422 0.14739 0.70708 0.47952
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7663
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.7470127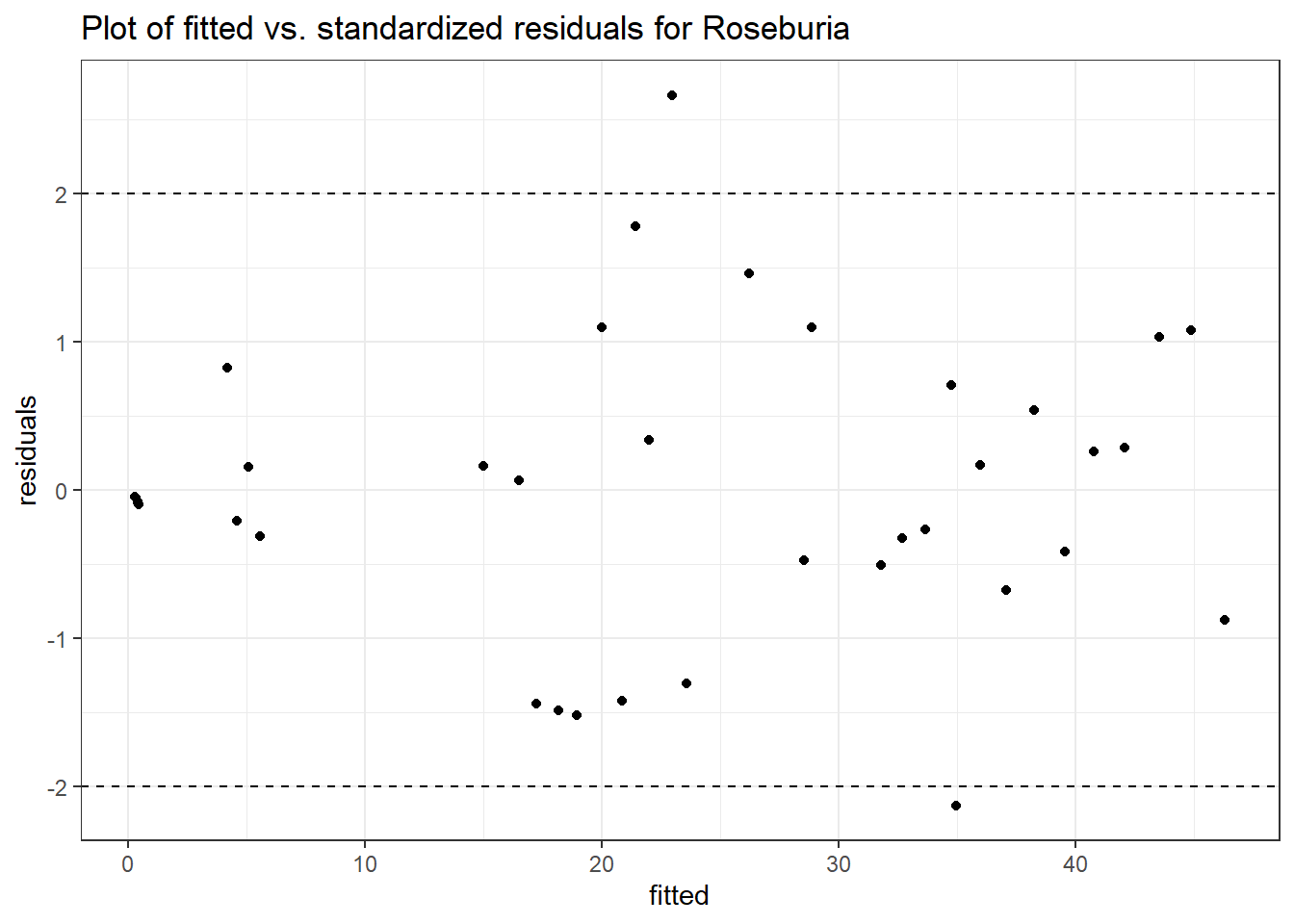

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 1.70580 0.75184 2.26884 0.02328
intB Roseburia 1.02812 1.07664 0.95493 0.33961
time Roseburia 0.09573 0.03045 3.14374 0.00167
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7184
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x00000000438186d8>
Link: poisson
Intraclass Correlation (ICC): 0.787515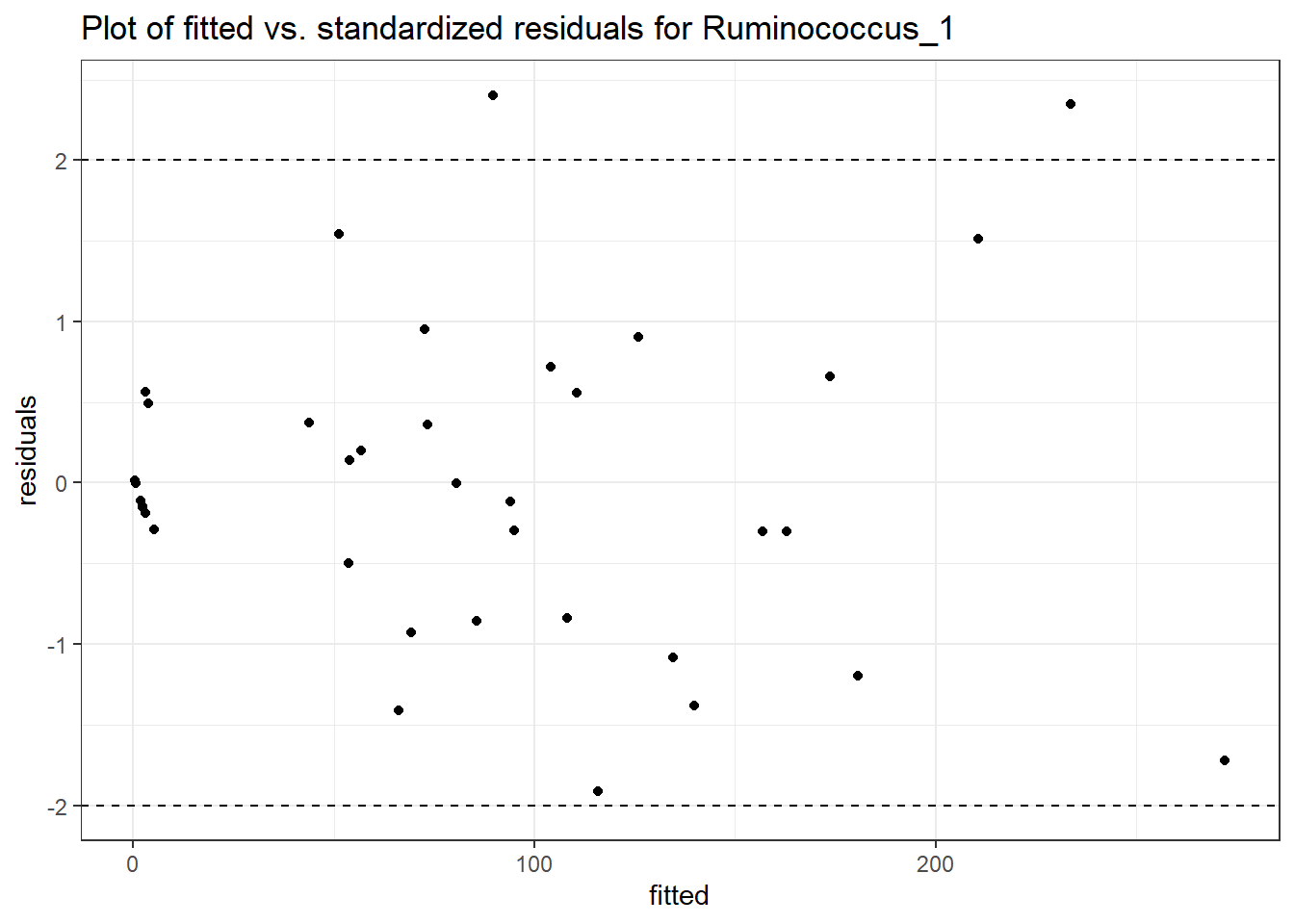
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 2.75494 0.80543 3.42046 0.00063
intB Ruminococcus_1 0.74558 1.18027 0.63171 0.52758
time Ruminococcus_1 0.25676 0.01598 16.06355 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.9252
Significance test not available for random effectsgenus.fit4b <- glmm_microbiome(mydata=microbiome_data,model.number=4,
taxa.level="Genus", taxa.subset = backNames,
link="negbinom",
model="1 + intB + time + (1|SubjectID)")
#########################################
#########################################
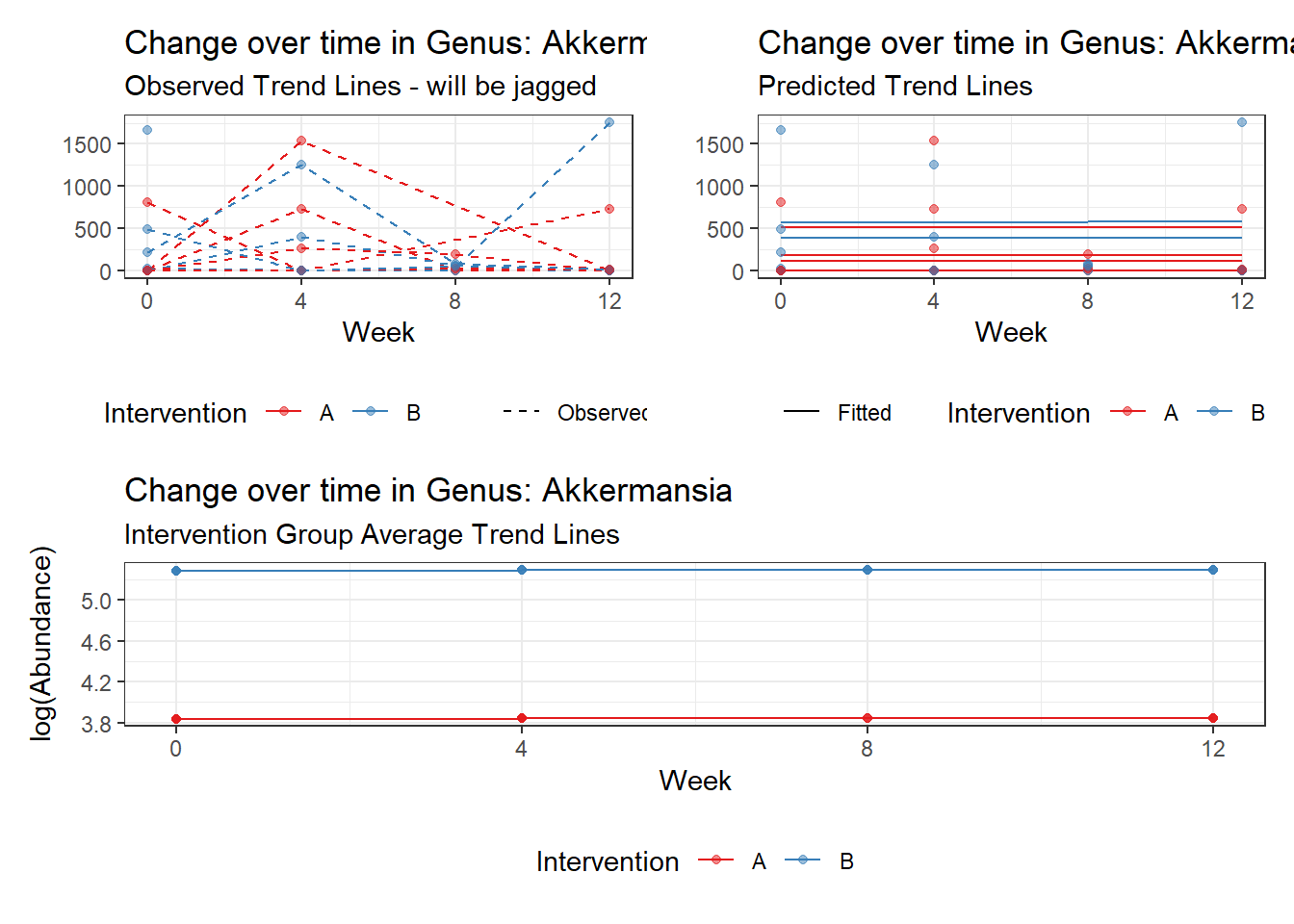
Model: Akkermansia ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced
Intraclass Correlation (ICC): 0.8511595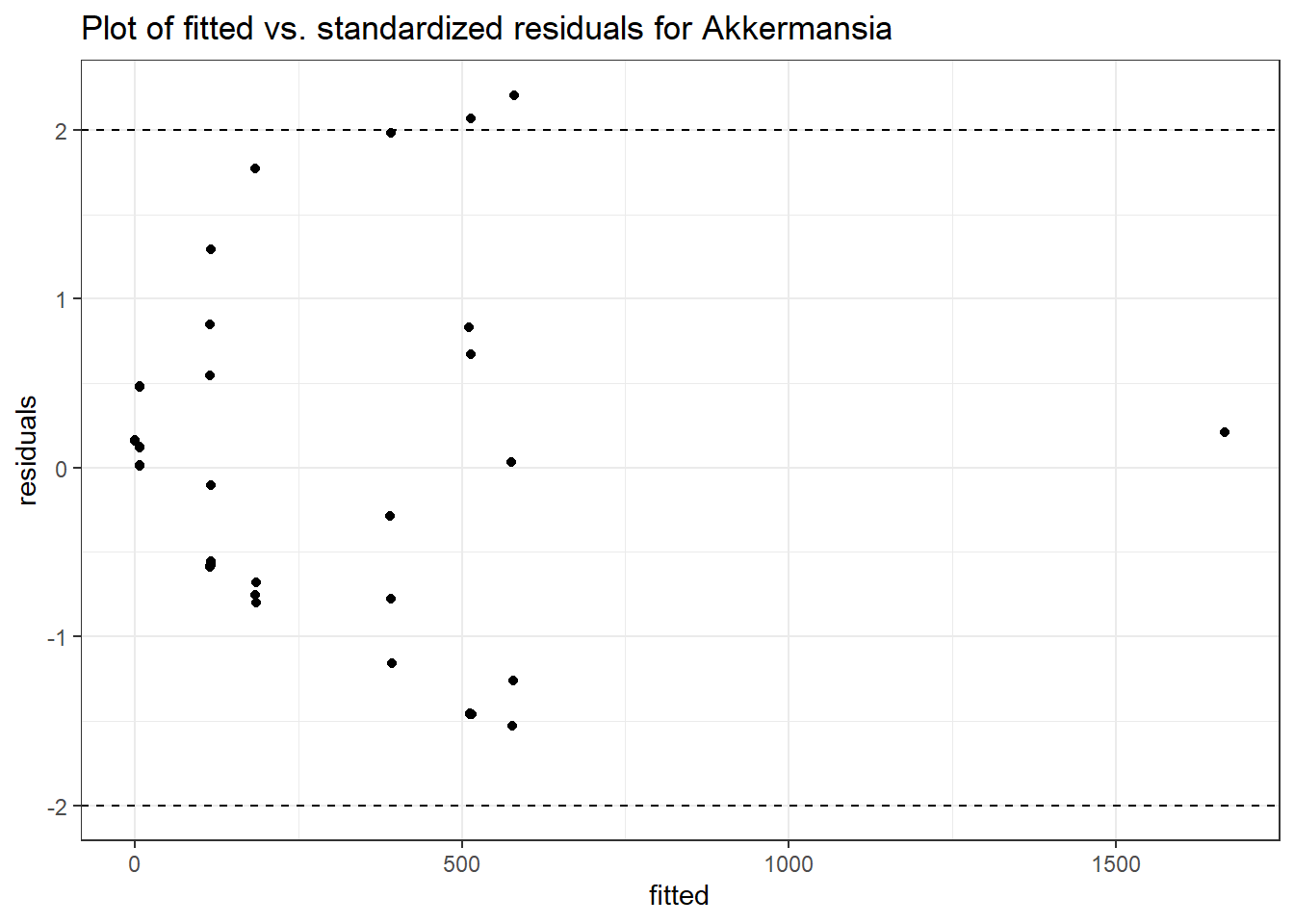
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 3.83782 0.99389 3.86142 0.00011
intB Akkermansia 1.45103 1.46178 0.99264 0.32088
time Akkermansia 0.00189 0.00938 0.20199 0.83993
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3914
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinom
Intraclass Correlation (ICC): 0.2150892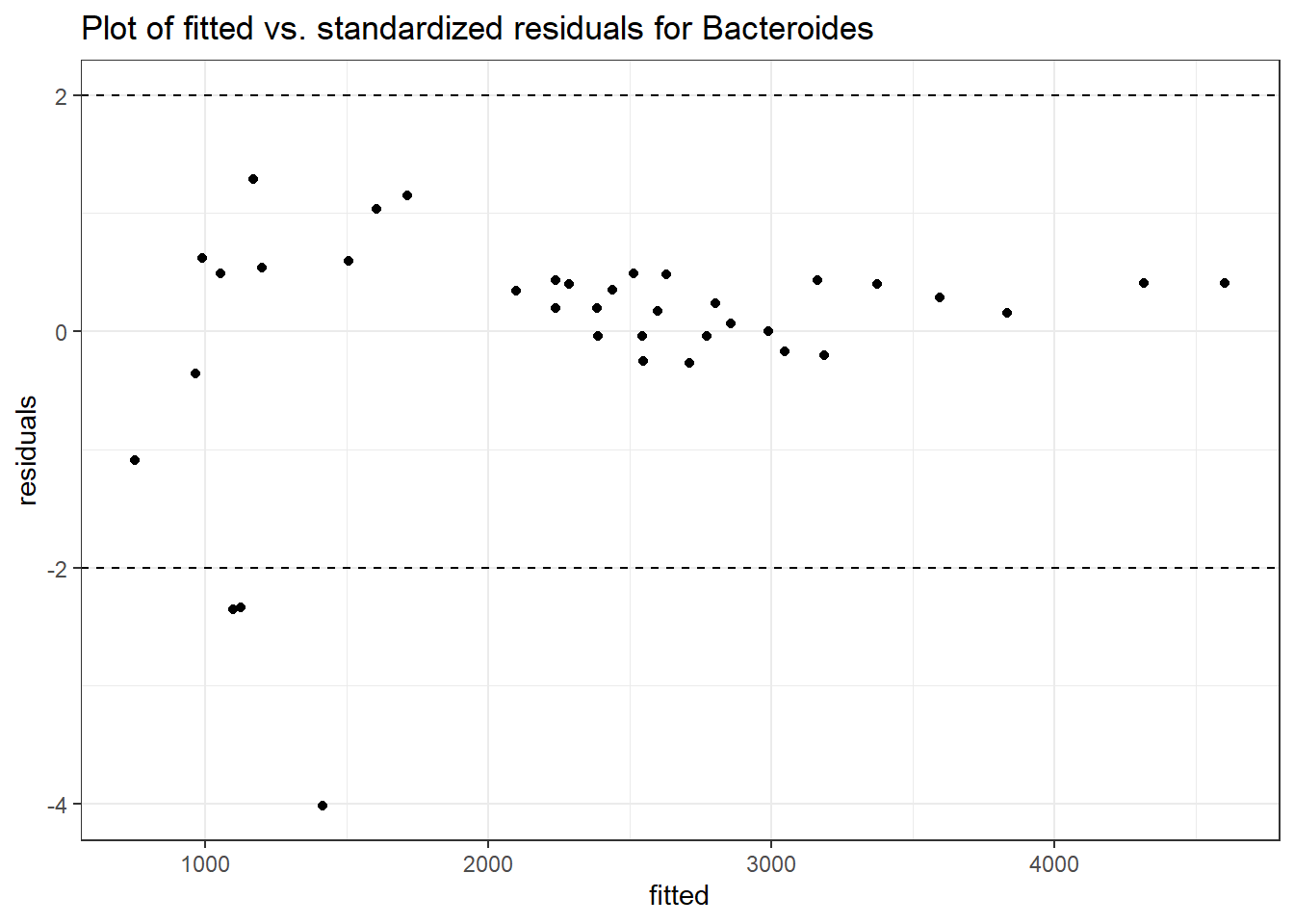

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.88980 0.23191 34.02123 0.00000
intB Bacteroides -0.42164 0.33682 -1.25181 0.21064
time Bacteroides -0.06413 0.04456 -1.43911 0.15012
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.52348
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinom
Intraclass Correlation (ICC): 0.28505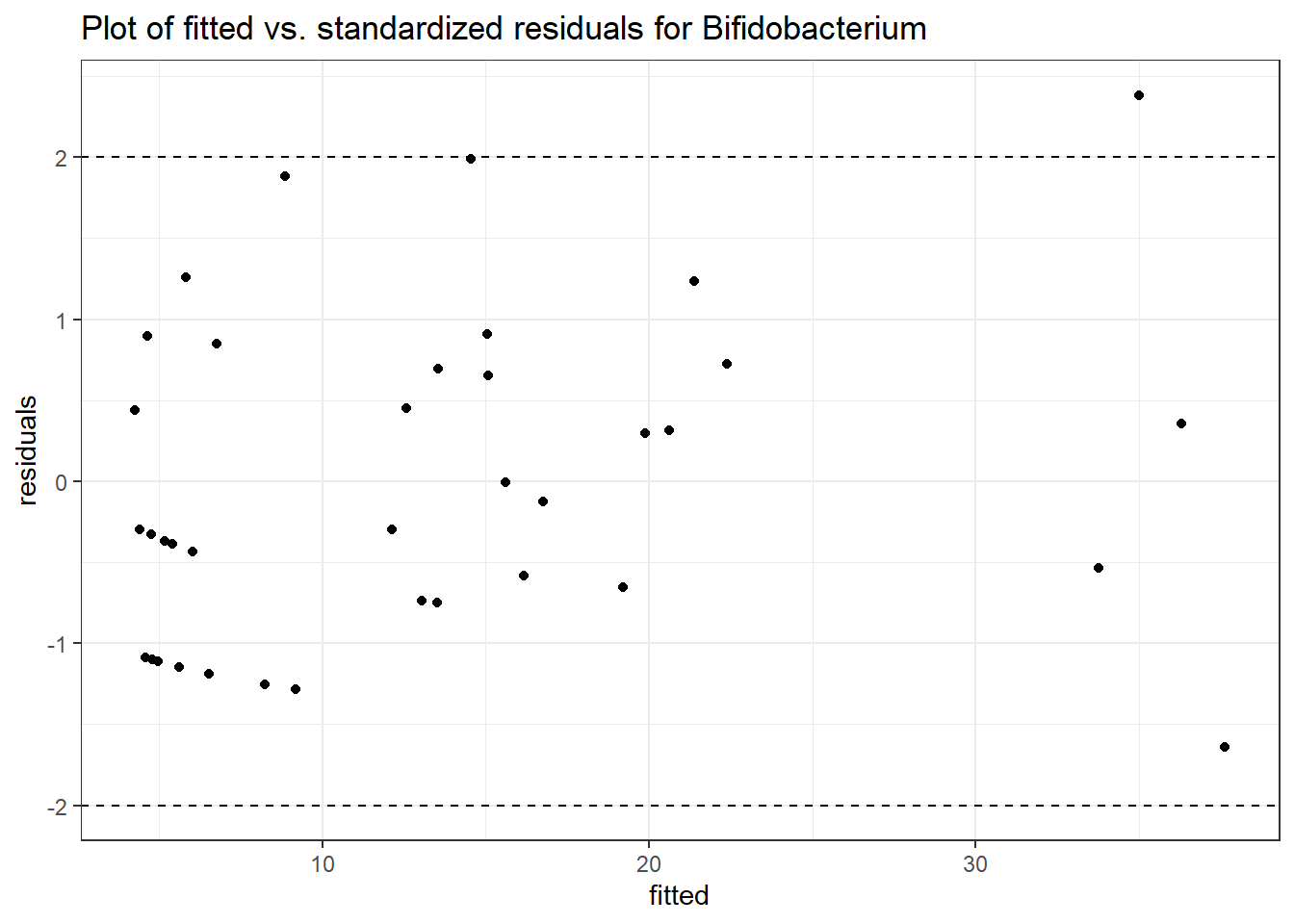
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 1.97131 0.62109 3.17396 0.00150
intB Bifidobacterium 1.07231 0.67115 1.59772 0.11010
time Bifidobacterium -0.03605 0.27971 -0.12889 0.89745
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.63143
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 1.740624e-13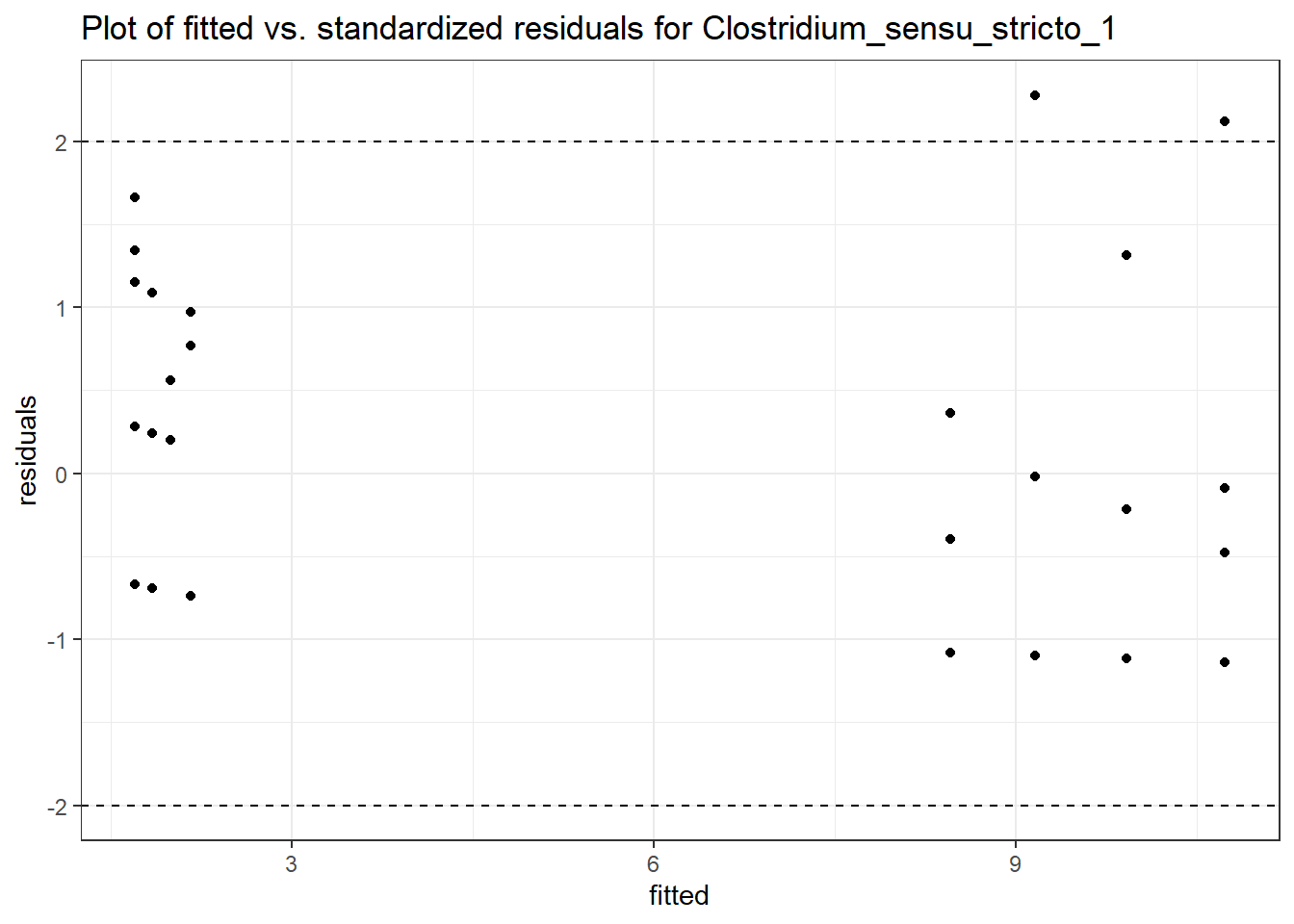

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.53305 0.53042 1.00496 0.31491
intB Clostridium_sensu_stricto_1 1.60221 0.67820 2.36243 0.01816
time Clostridium_sensu_stricto_1 0.07943 0.29638 0.26800 0.78870
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.1721e-07
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinom
Intraclass Correlation (ICC): 0.01261025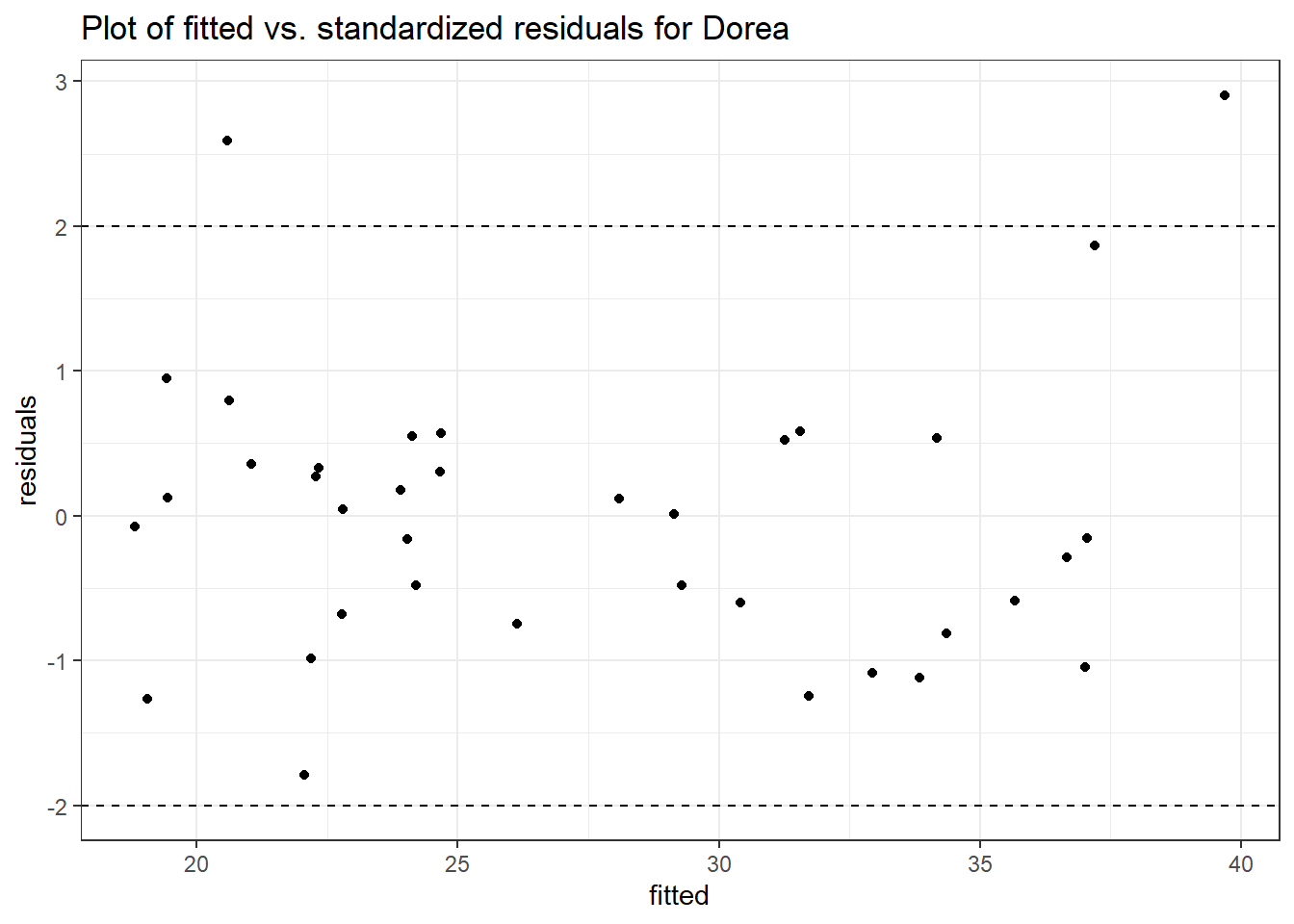
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.20241 0.22975 13.93840 0.00000
intB Dorea 0.41654 0.24705 1.68605 0.09179
time Dorea -0.07957 0.09591 -0.82966 0.40673
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.11301
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinomboundary (singular) fit: see ?isSingular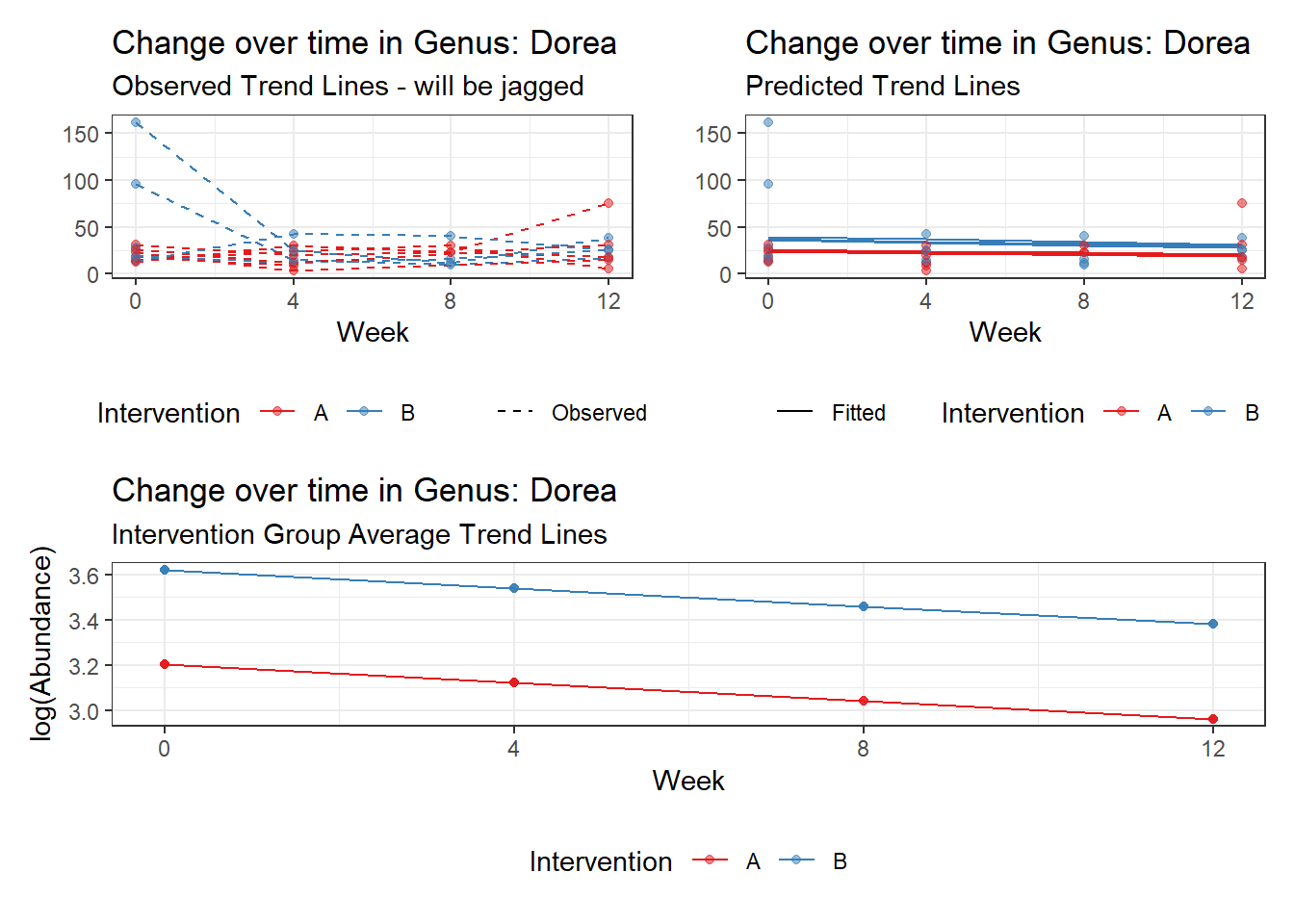
Intraclass Correlation (ICC): 2.407996e-11
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.81325 0.30471 22.35944 0.00000
intB Faecalibacterium -0.25763 0.31618 -0.81483 0.41517
time Faecalibacterium -0.04323 0.15285 -0.28282 0.77731
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.9071e-06
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reached
Warning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, : NaNs
produced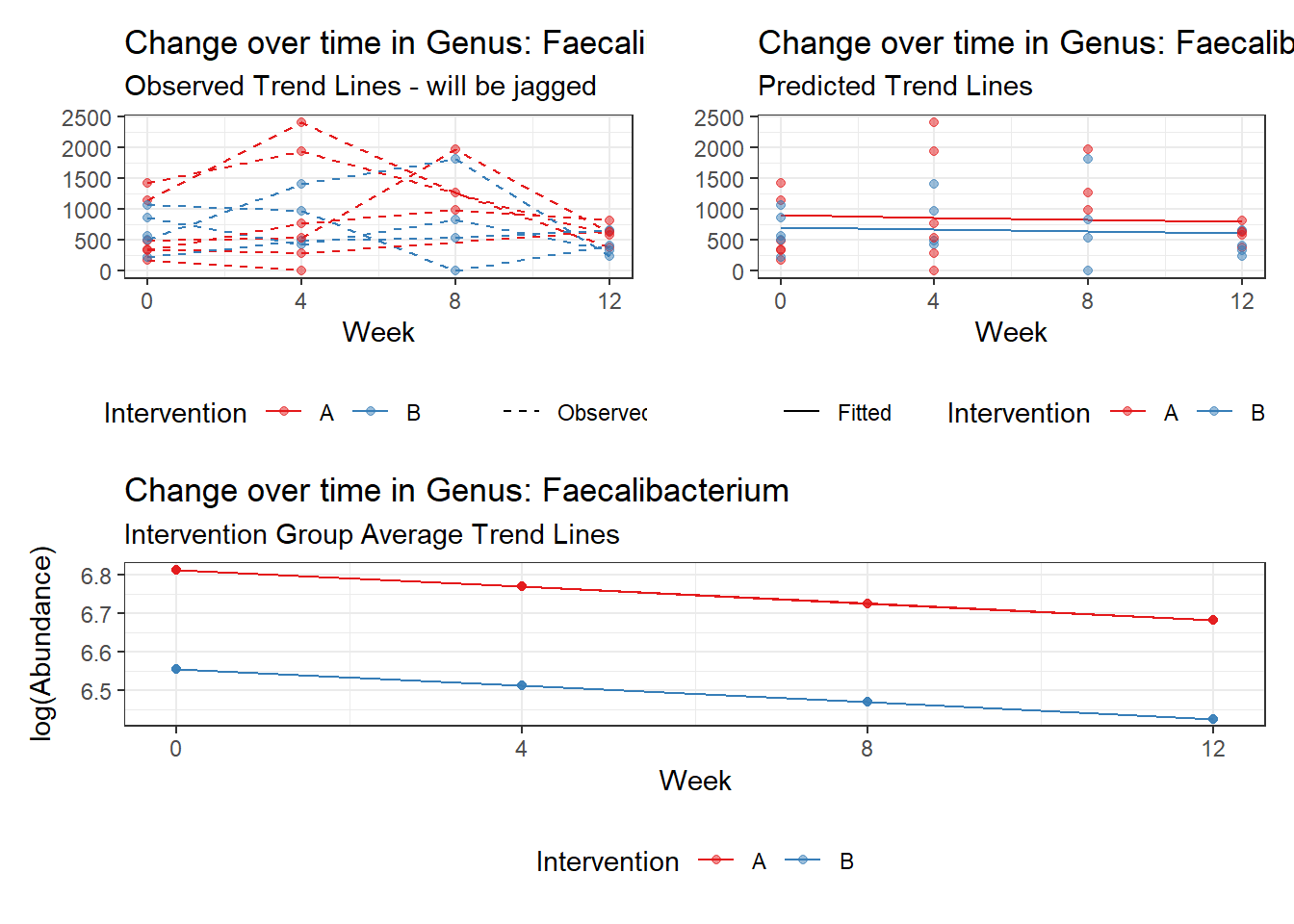
Intraclass Correlation (ICC): 0.2926728
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.14709 0.26534 15.62952 0.00000
intB Lachnospira -0.77188 0.39534 -1.95242 0.05089
time Lachnospira 0.11969 0.01779 6.72863 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.64325
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinomboundary (singular) fit: see ?isSingular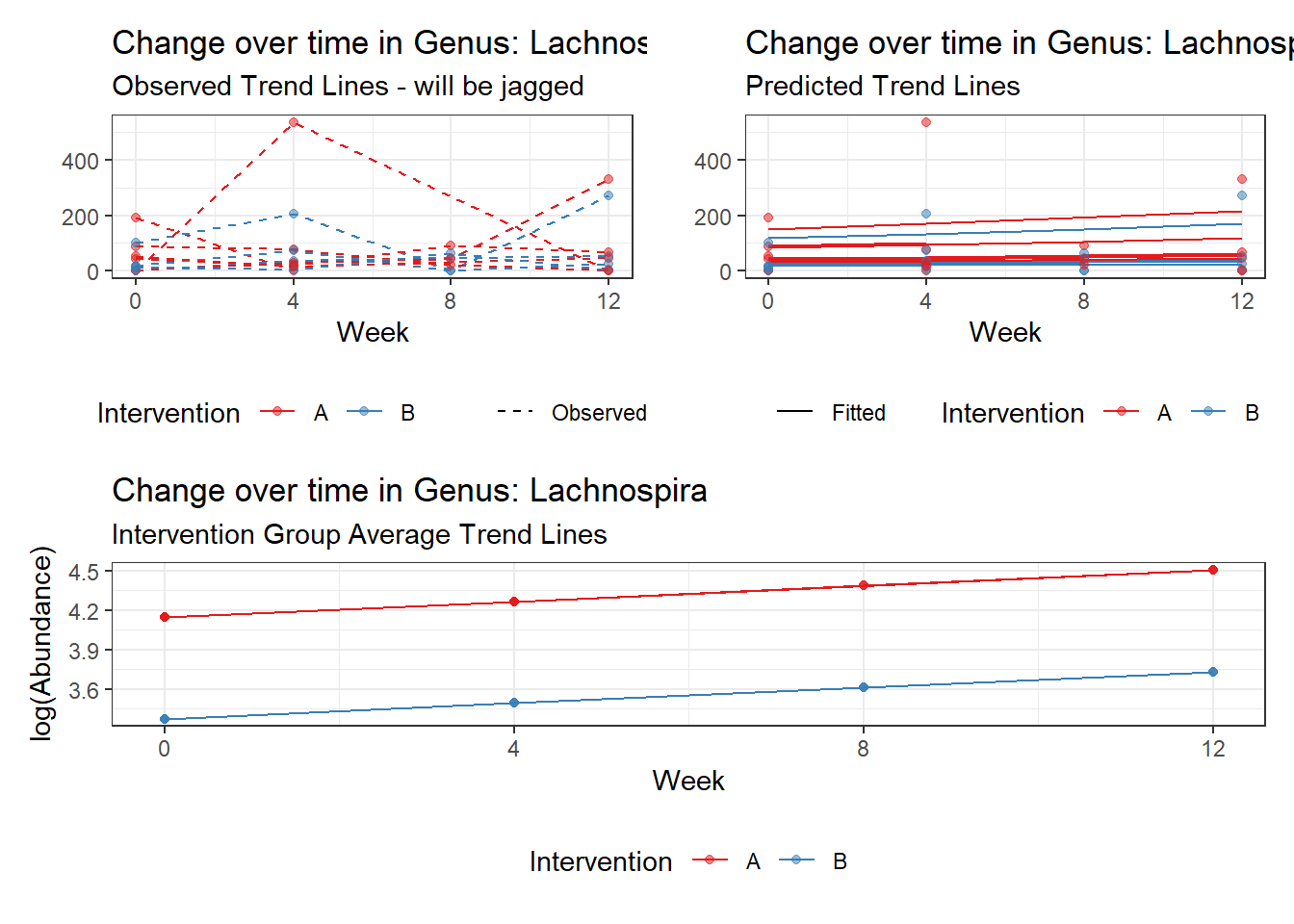
Intraclass Correlation (ICC): 1.246477e-11
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus 2.84309 1.14397 2.48529 0.01294
intB Lactobacillus -2.99323 1.60624 -1.86349 0.06239
time Lactobacillus -1.01146 0.88972 -1.13683 0.25561
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.5305e-06
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 6.419327e-14
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -0.12759 1.09034 -0.11702 0.90684
intB Prevotella 0.41050 1.09242 0.37577 0.70709
time Prevotella -0.02225 0.51214 -0.04344 0.96535
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.5336e-07
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinomboundary (singular) fit: see ?isSingular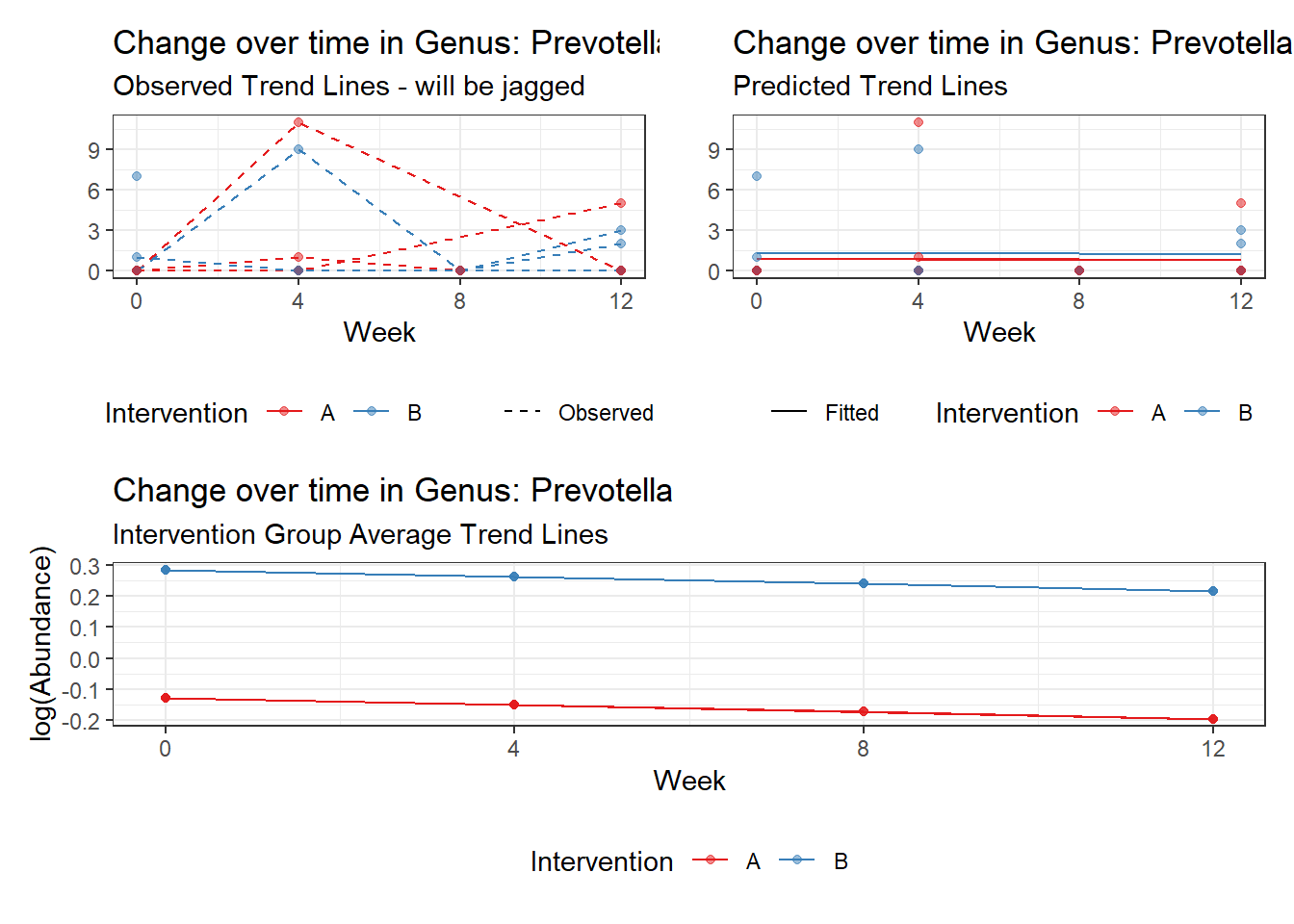
Intraclass Correlation (ICC): 1.166882e-15
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 2.96005 0.43479 6.80802 0.00000
intB Roseburia -0.04095 0.49643 -0.08250 0.93425
time Roseburia 0.13686 0.20483 0.66814 0.50404
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.416e-08
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003f31fc98>
Link: negbinomboundary (singular) fit: see ?isSingular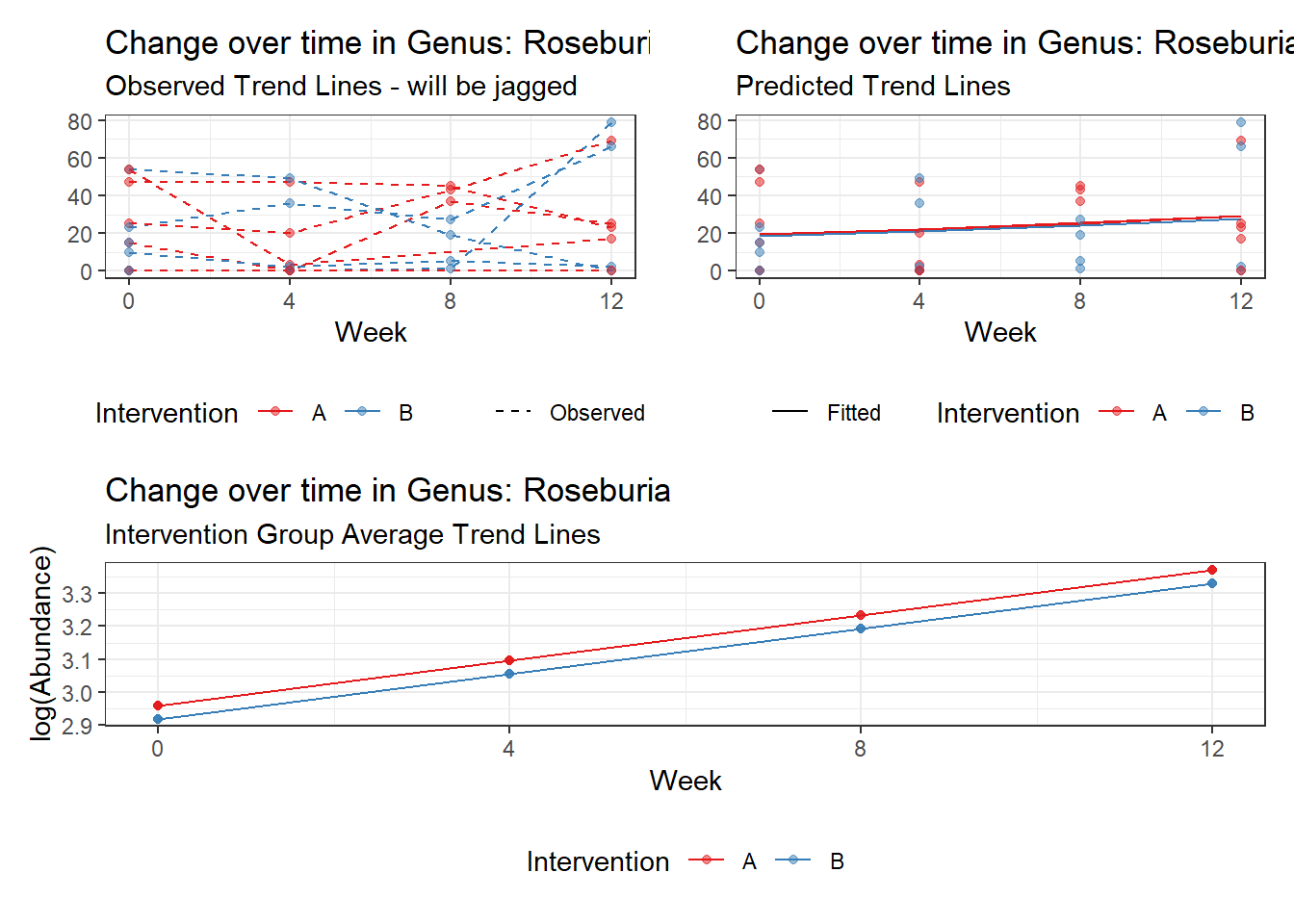
Intraclass Correlation (ICC): 5.222402e-11
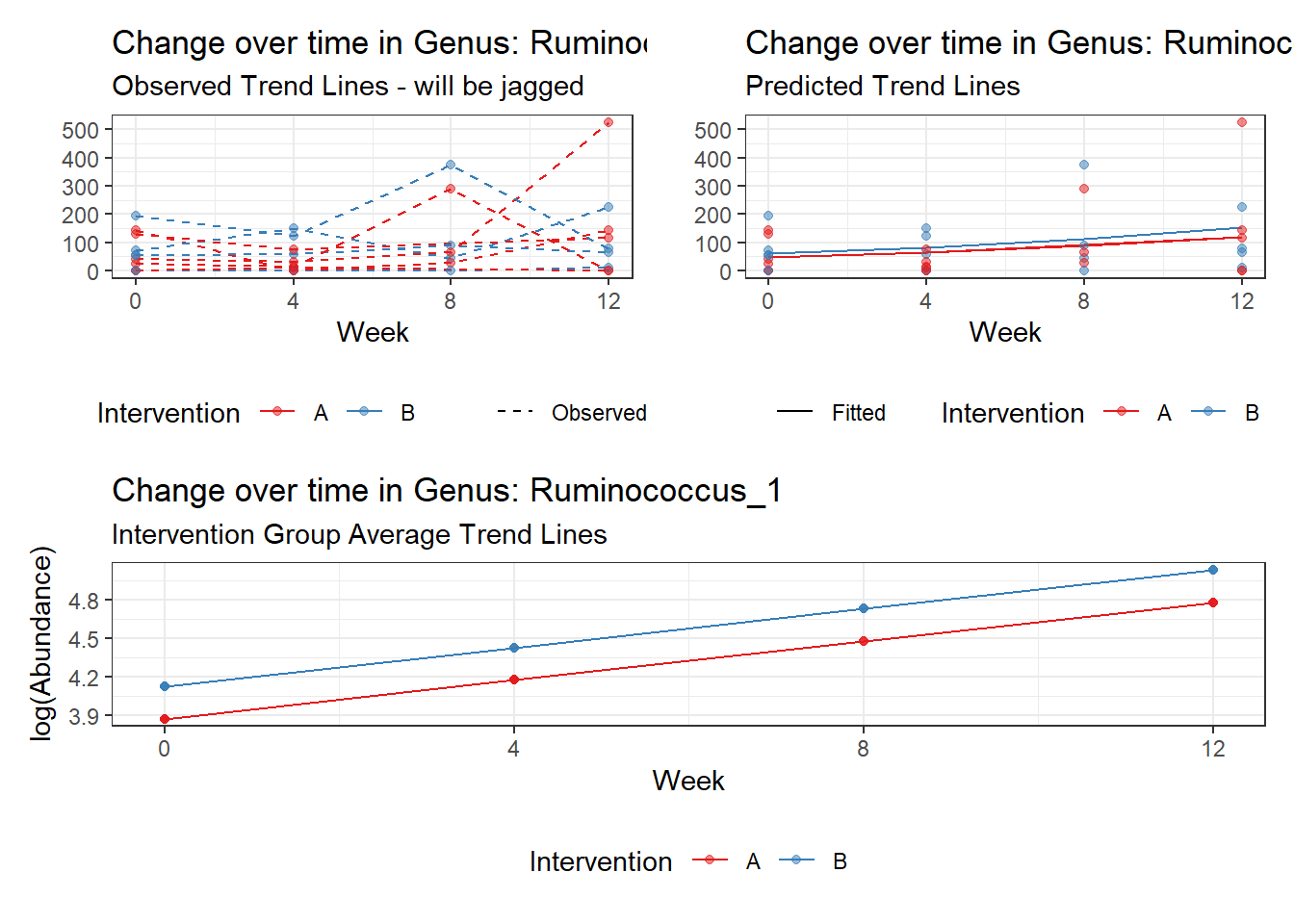
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 3.87419 0.51488 7.52452 0.00000
intB Ruminococcus_1 0.25325 0.55585 0.45561 0.64867
time Ruminococcus_1 0.30159 0.23139 1.30338 0.19244
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 7.2266e-06
Significance test not available for random effectsControlling for the effect of time, the intervention resulted in a significant increase in abundance of Genuss:
Other interesting but not significant results. The effect of the intervention was in the opposite direction (positive) compared to the effect of time (negative) is Genus:
Covariate(s): Time by intervention interaction
Here, the model tests whether the effect of the intervention is constant across time or does the effect of the intervention get stronger (or weaker) at time progress. And remember, time is coded 0, 1, 2, 3, so a 1 unit increase in time corresponds to 1 month or 4 weeks of the intervention.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Intervention)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
It could be that the intervention is directly related to time so included a average effect of time through the main effect effect is just adding more parameters and is not informative. If the effect of the intervention is still essentially zero, we will re-estimate the model without the main effect.
genus.fit5 <- glmm_microbiome(mydata=microbiome_data,model.number=5,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + intB + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.8554283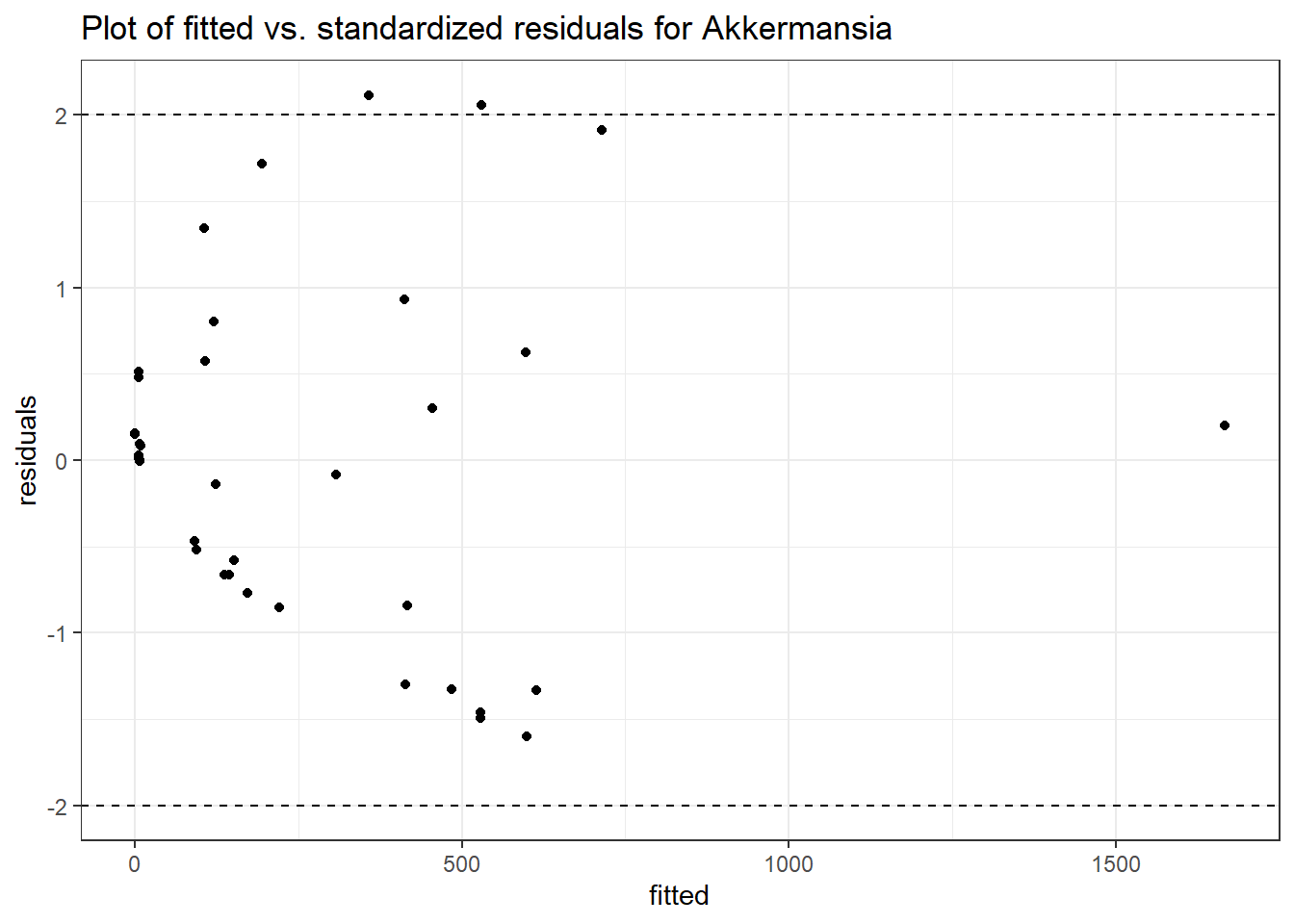

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 3.98707 1.00988 3.94807 0.00008
intB Akkermansia 1.11249 1.48508 0.74911 0.45379
time Akkermansia -0.12308 0.01296 -9.49425 0.00000
intB:time Akkermansia 0.27329 0.01883 14.51628 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4325
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.2604204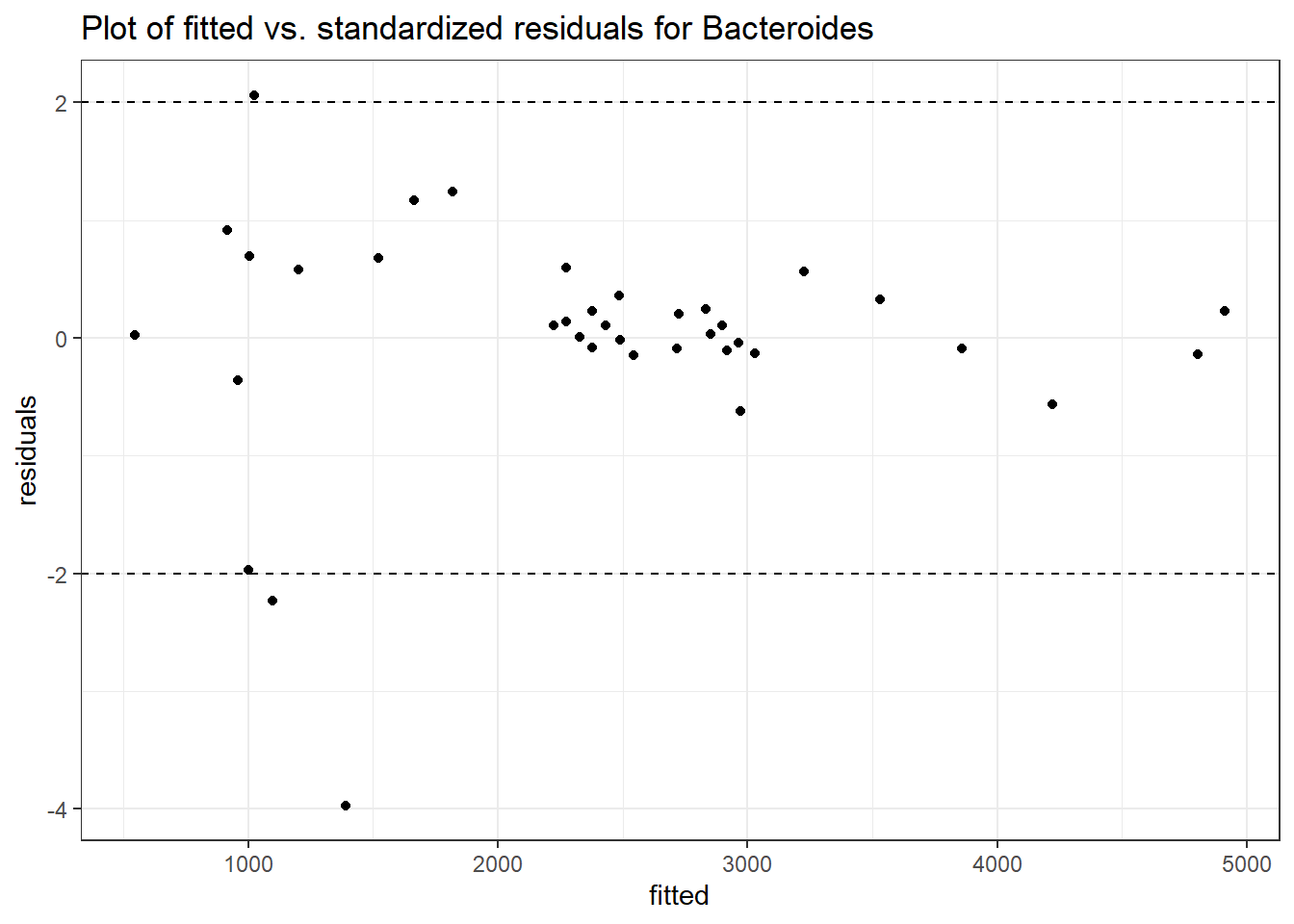

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.84043 0.24236 32.35055 0.00000
intB Bacteroides -0.39199 0.35956 -1.09019 0.27563
time Bacteroides -0.02259 0.00413 -5.46511 0.00000
intB:time Bacteroides -0.06692 0.00629 -10.64403 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.5934
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.5043246

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 1.90771 0.43177 4.41834 0.00001
intB Bifidobacterium 0.81194 0.63108 1.28660 0.19824
time Bifidobacterium -0.11367 0.06204 -1.83210 0.06694
intB:time Bifidobacterium 0.25013 0.07635 3.27588 0.00105
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0087
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + intB + time + intB:time + (1 |
SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.6125261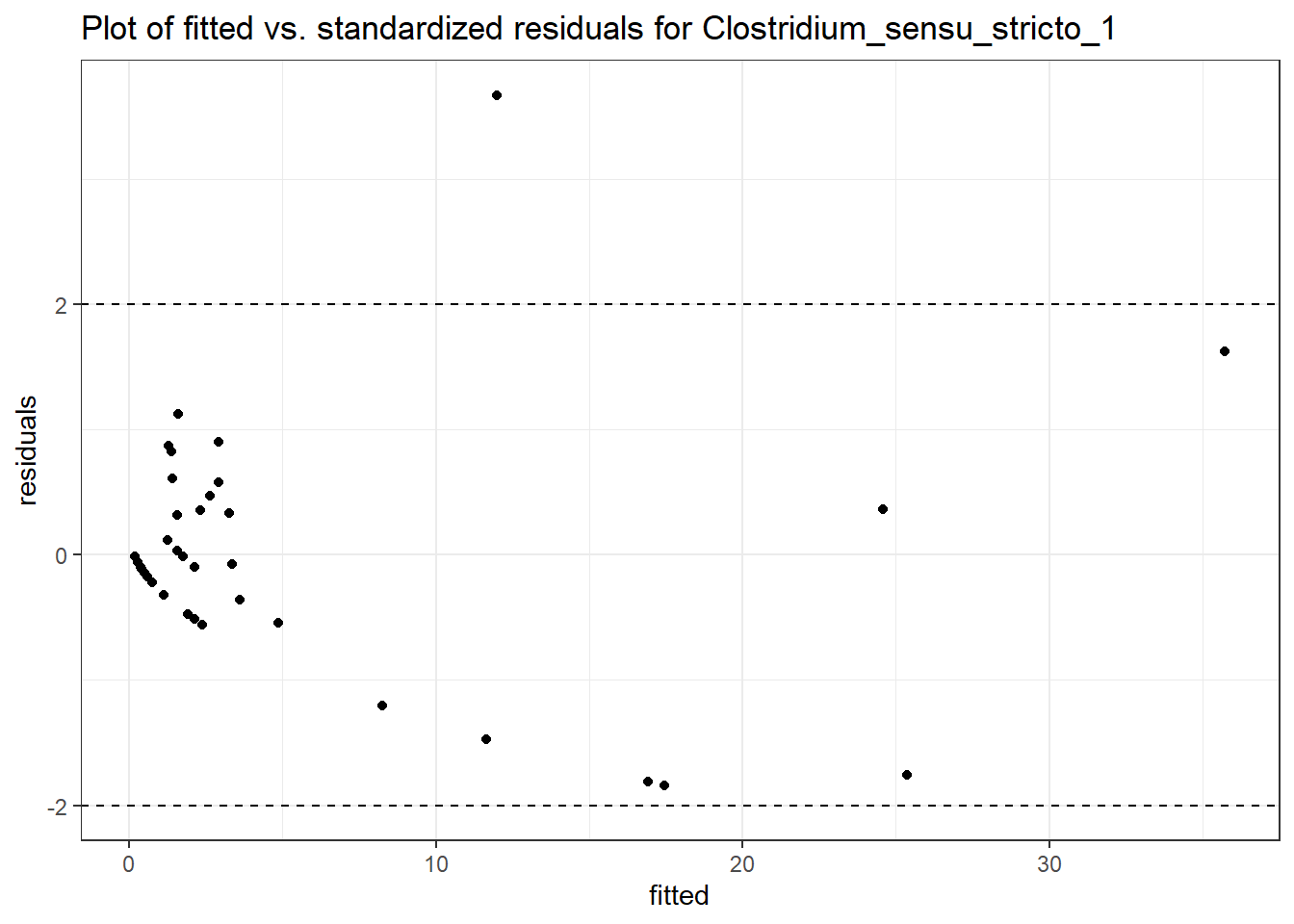
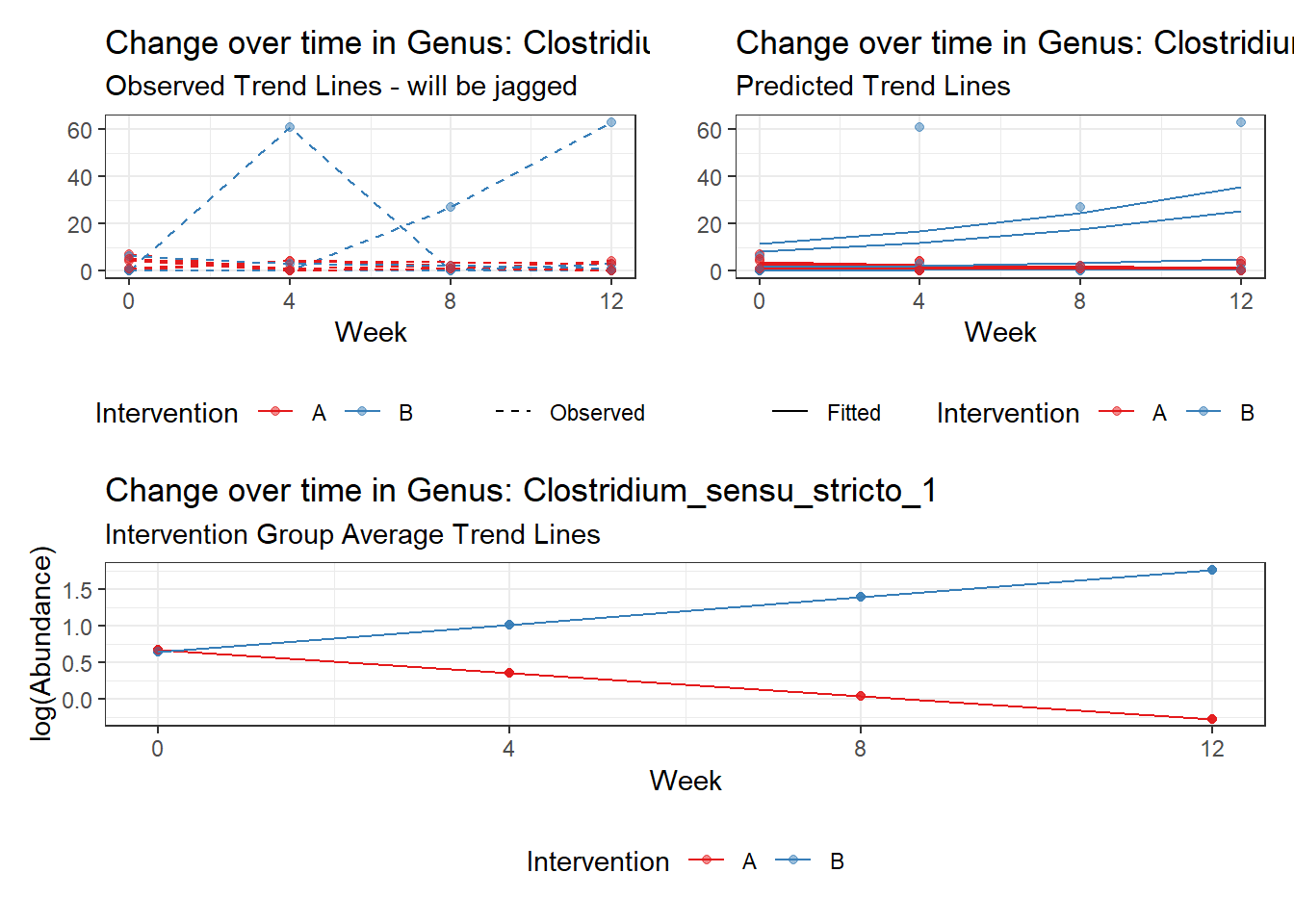
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.67852 0.57409 1.18192 0.23724
intB Clostridium_sensu_stricto_1 -0.03620 0.84221 -0.04298 0.96571
time Clostridium_sensu_stricto_1 -0.31592 0.14946 -2.11374 0.03454
intB:time Clostridium_sensu_stricto_1 0.69010 0.16628 4.15012 0.00003
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.2573
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.1217556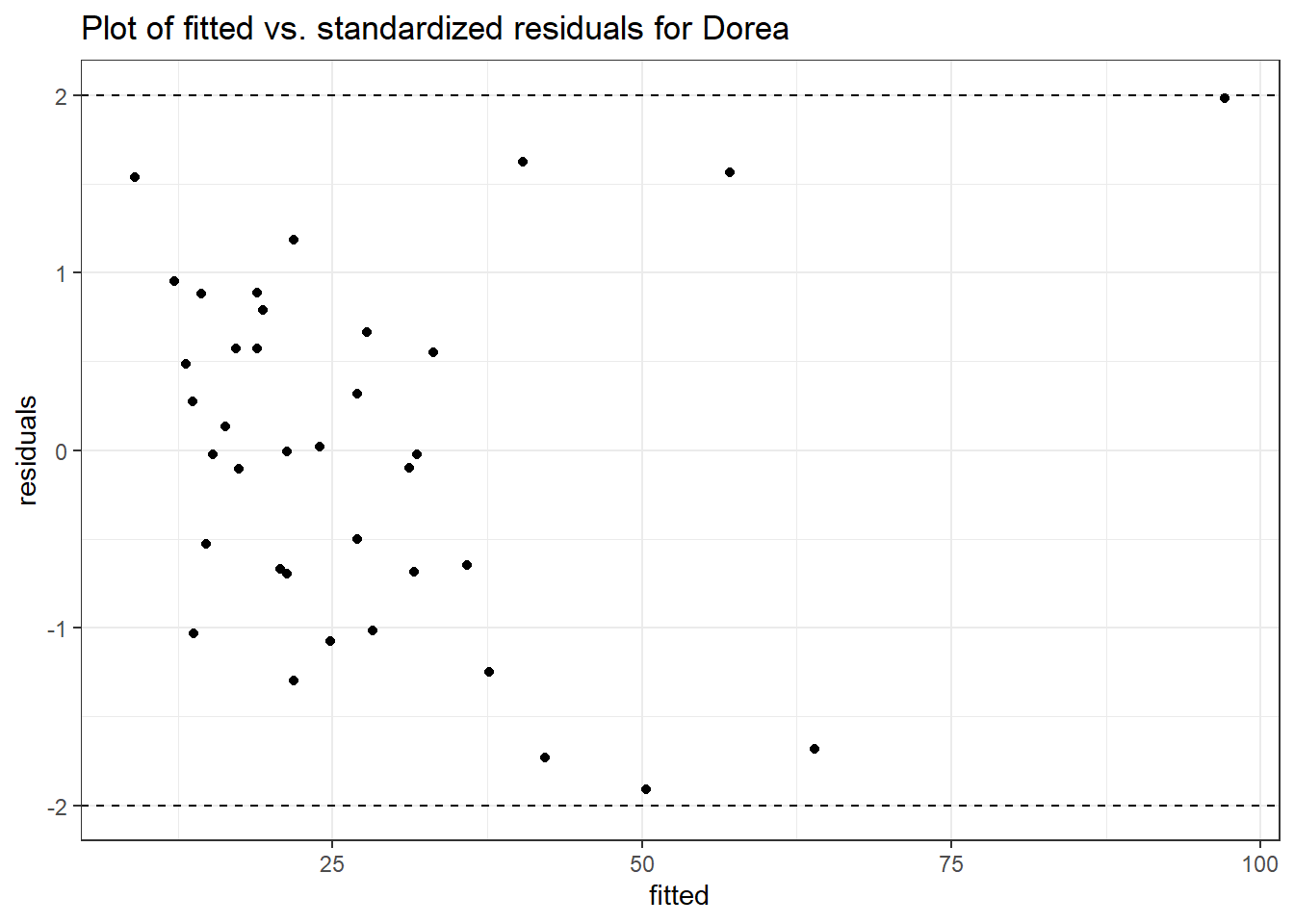
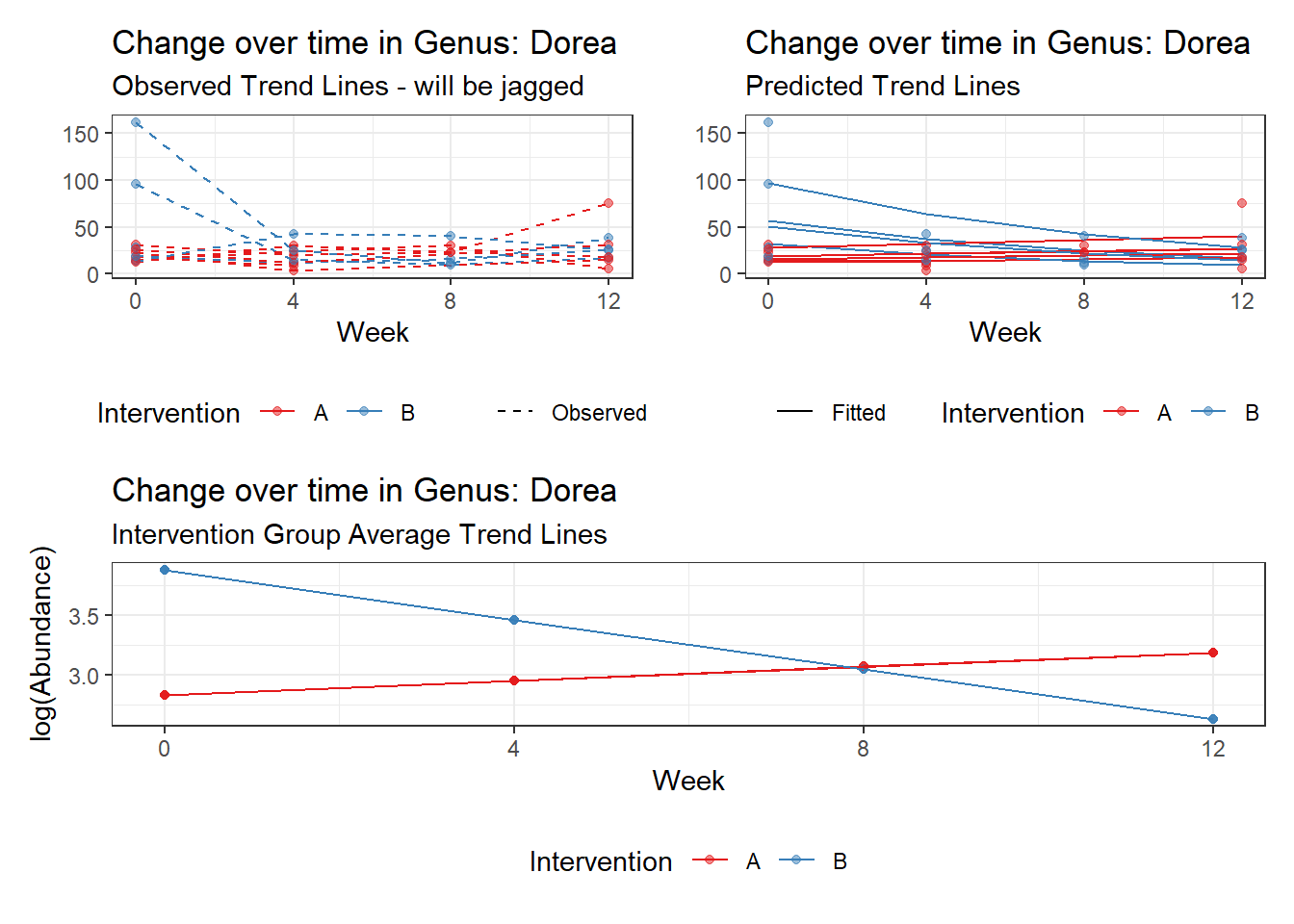
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 2.83040 0.17212 16.44422 0.00000
intB Dorea 1.05100 0.24688 4.25719 0.00002
time Dorea 0.11893 0.04204 2.82883 0.00467
intB:time Dorea -0.53647 0.05822 -9.21429 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.37234
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.3419755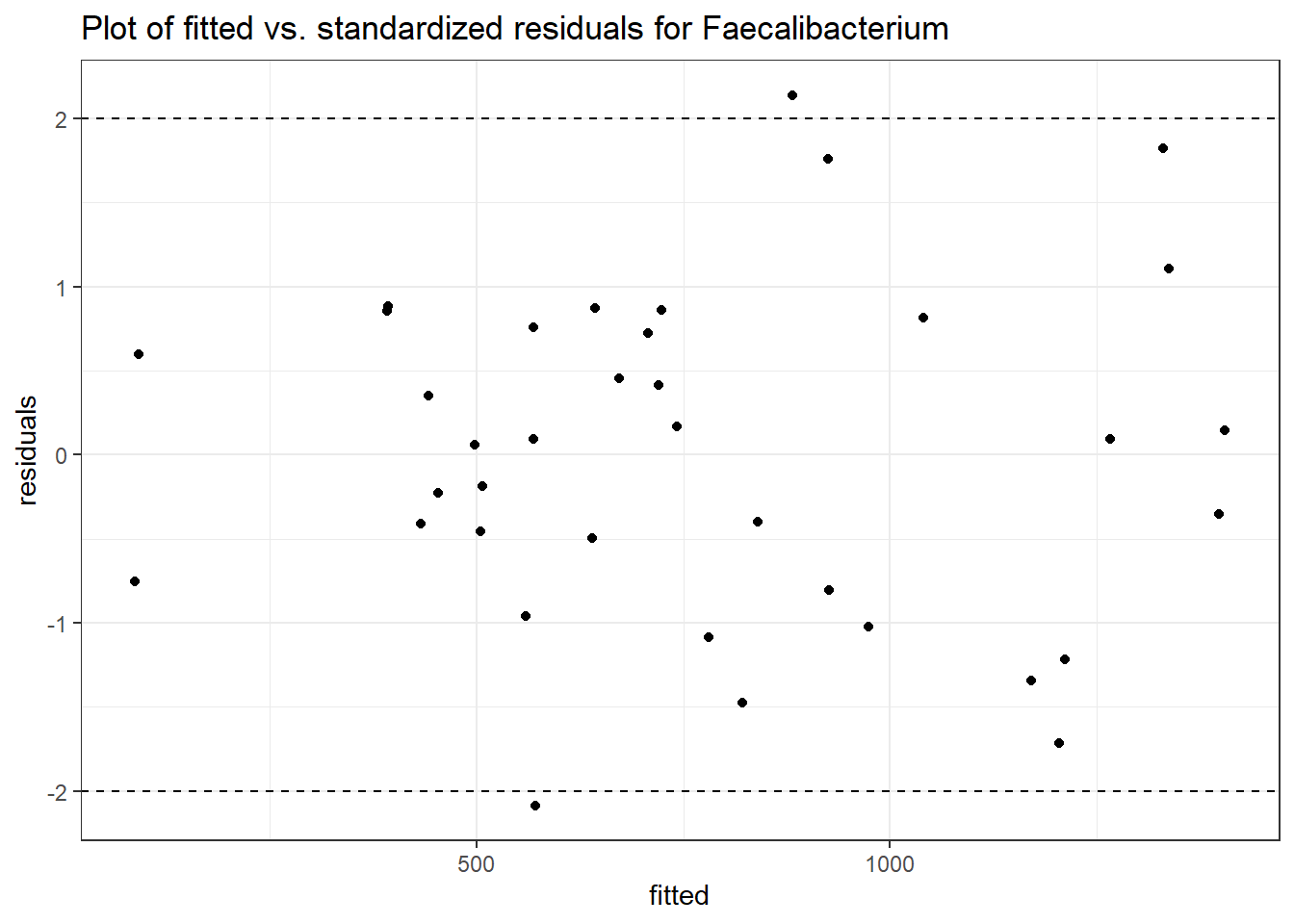
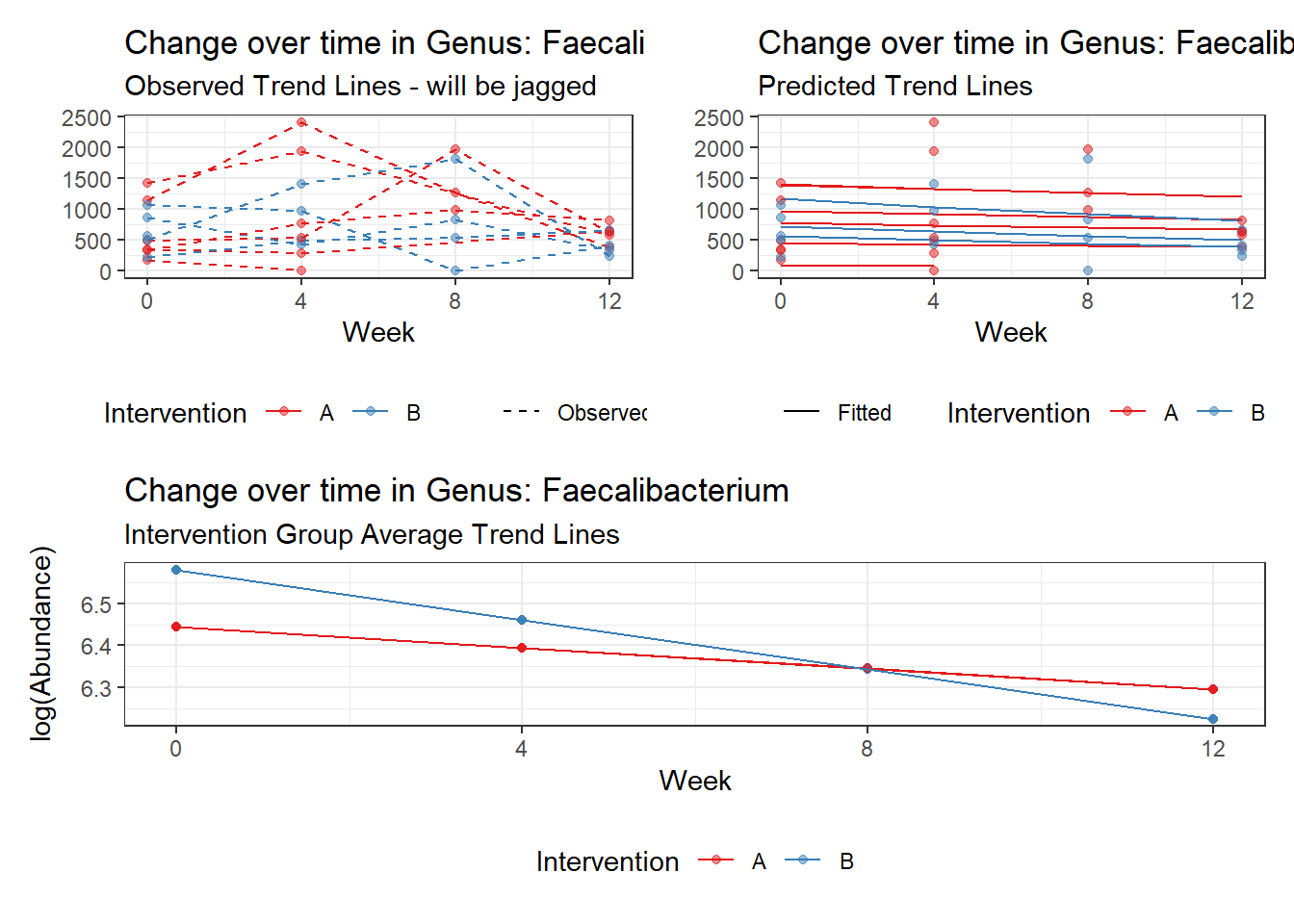
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.44356 0.29477 21.85997 0.00000
intB Faecalibacterium 0.13592 0.43708 0.31097 0.75582
time Faecalibacterium -0.04934 0.00665 -7.41566 0.00000
intB:time Faecalibacterium -0.06871 0.01094 -6.28028 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.7209
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.2827835
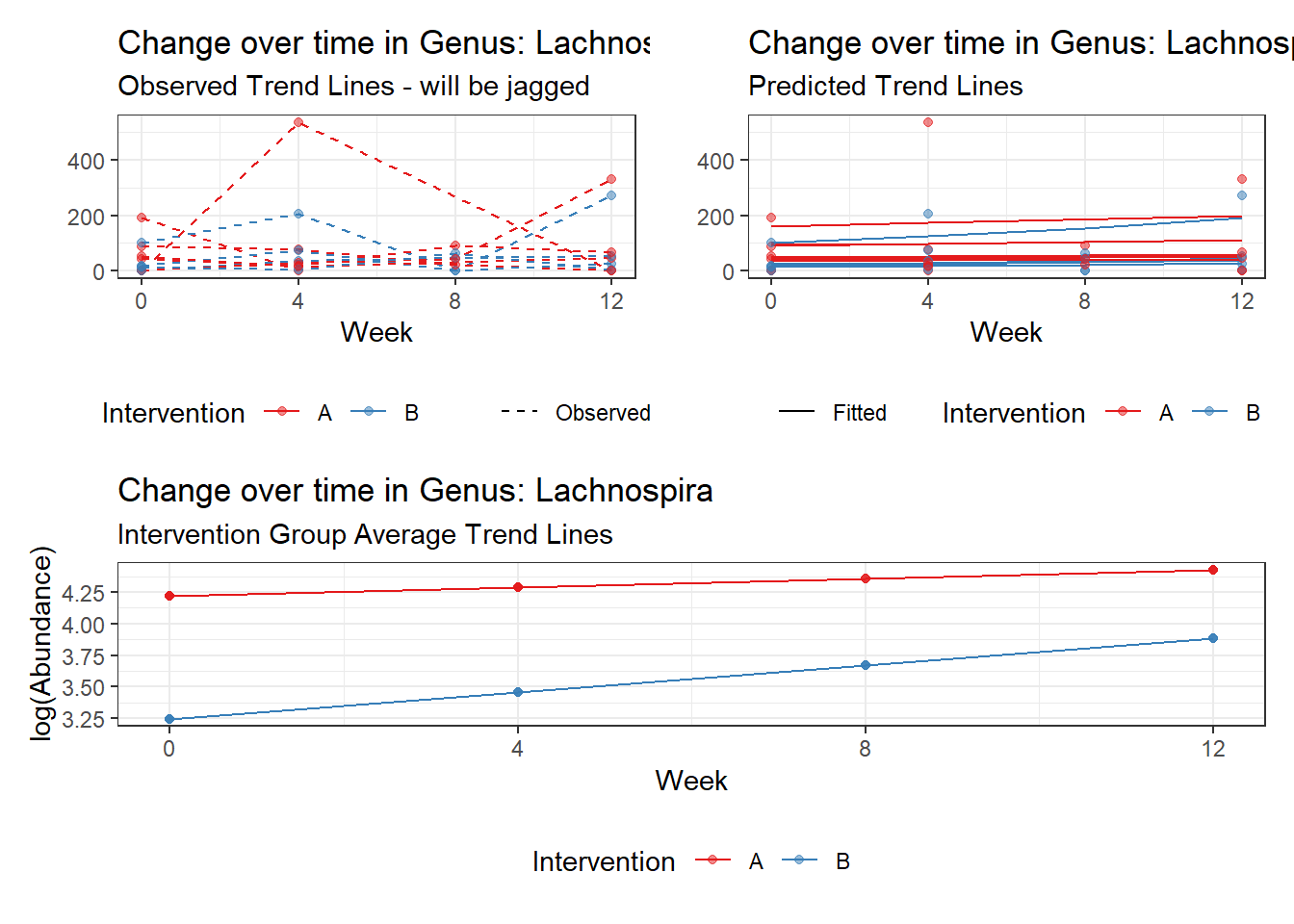
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.21709 0.25956 16.24679 0.00000
intB Lachnospira -0.96898 0.38947 -2.48796 0.01285
time Lachnospira 0.06924 0.02182 3.17236 0.00151
intB:time Lachnospira 0.14208 0.03716 3.82321 0.00013
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.62792
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.8445182

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus 0.09042 1.12244 0.08056 0.93579
intB Lactobacillus -3.25415 1.88762 -1.72395 0.08472
time Lactobacillus -0.98633 0.09739 -10.12789 0.00000
intB:time Lactobacillus 1.13671 0.57097 1.99082 0.04650
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3306
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.7506264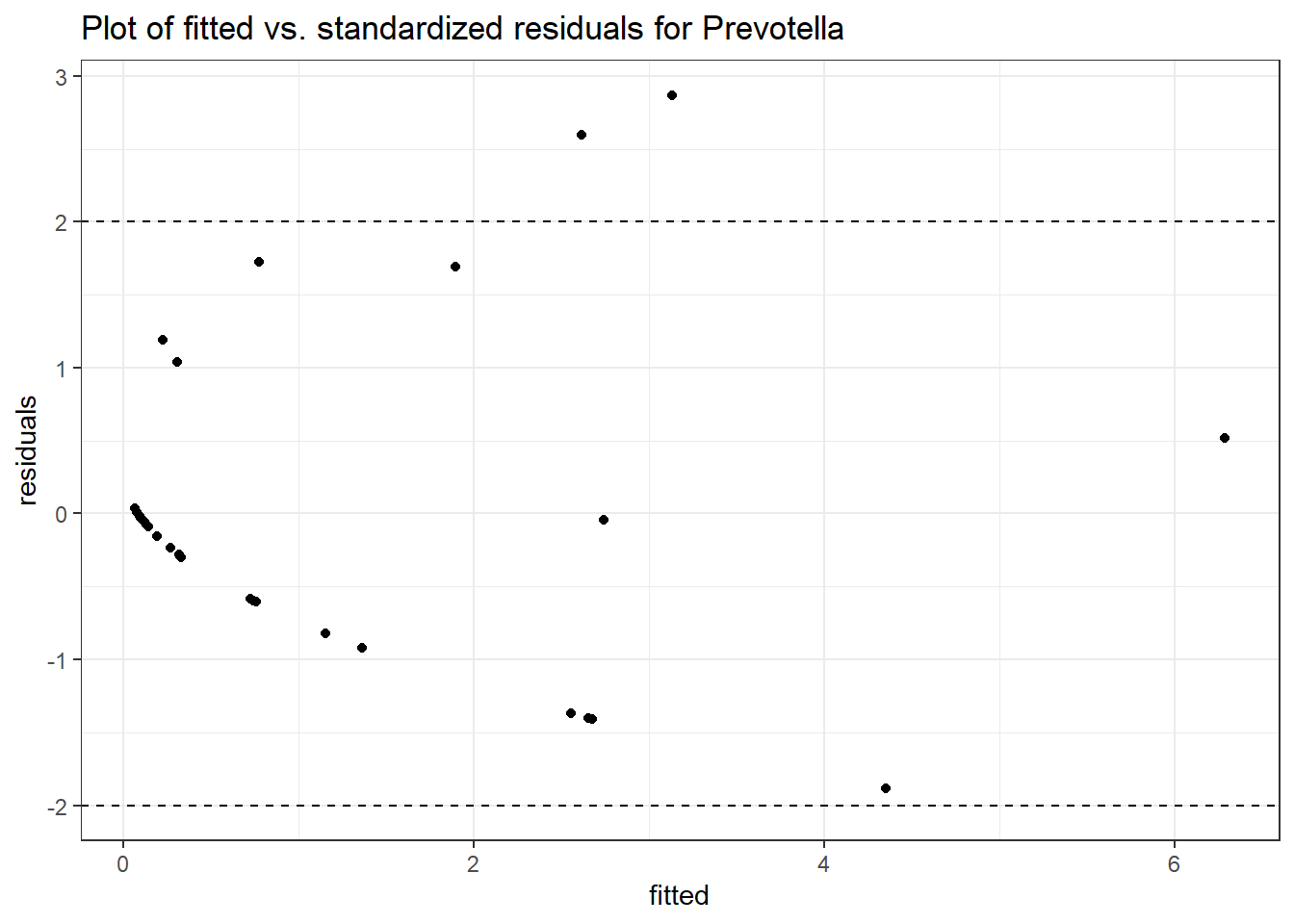
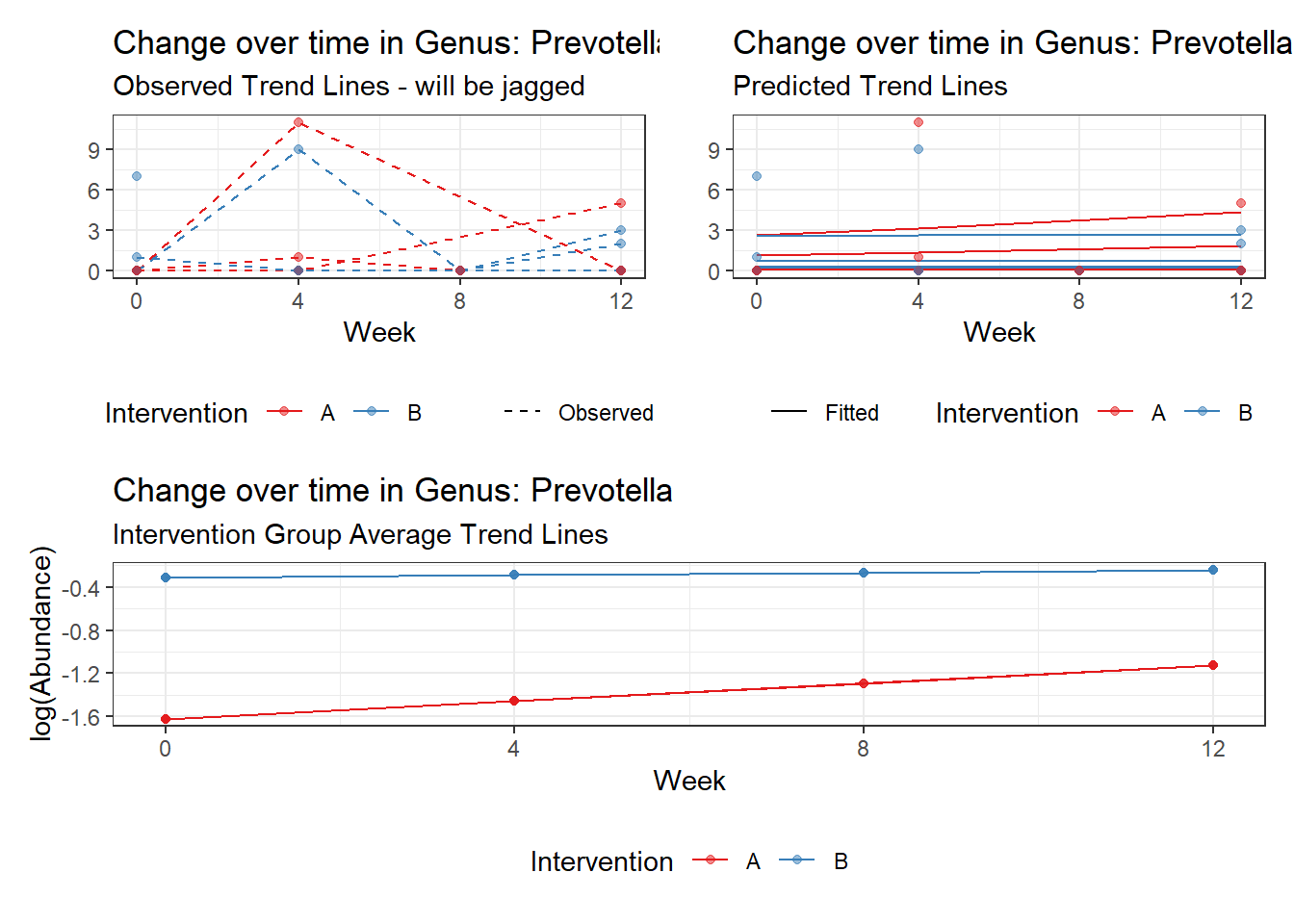
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.61959 1.00125 -1.61758 0.10575
intB Prevotella 1.30606 1.28765 1.01430 0.31044
time Prevotella 0.16454 0.19441 0.84635 0.39736
intB:time Prevotella -0.14091 0.29729 -0.47399 0.63551
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7349
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.7493791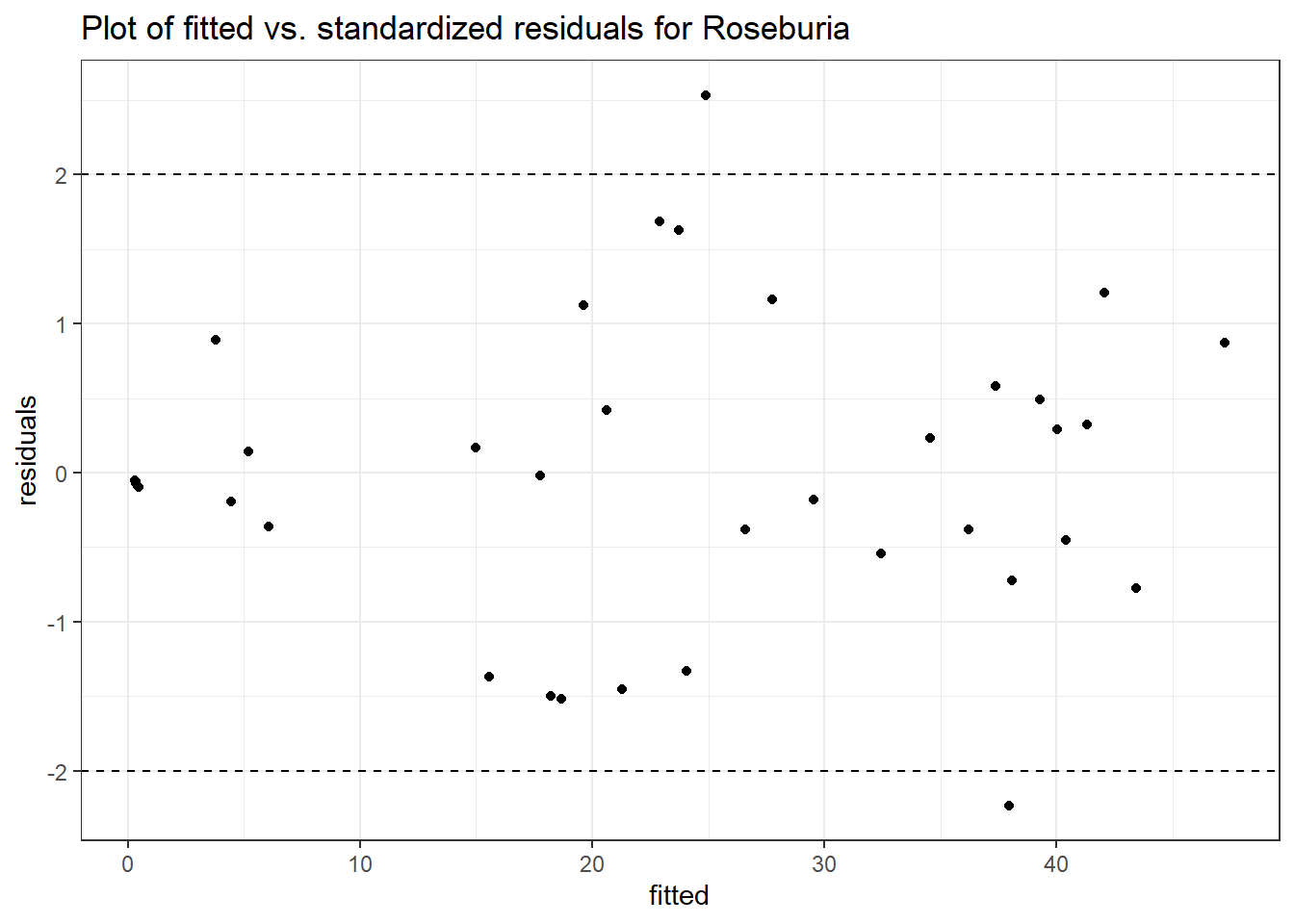

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 1.76595 0.75684 2.33331 0.01963
intB Roseburia 0.88703 1.08624 0.81661 0.41415
time Roseburia 0.04983 0.04036 1.23463 0.21697
intB:time Roseburia 0.10671 0.06167 1.73030 0.08358
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7292
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004d643c48>
Link: poisson
Intraclass Correlation (ICC): 0.779599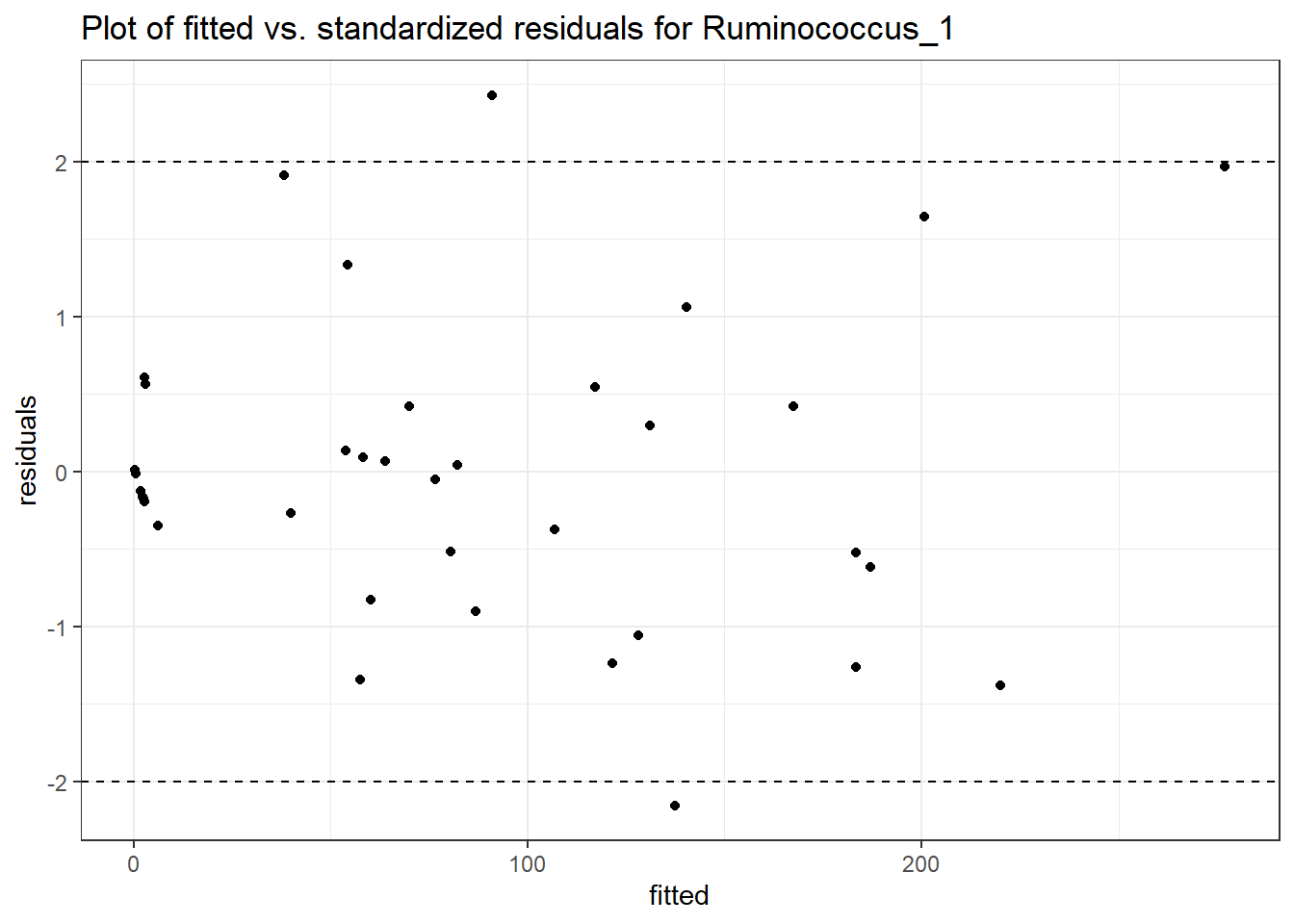

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 2.49673 0.78823 3.16752 0.00154
intB Ruminococcus_1 1.23204 1.15470 1.06699 0.28598
time Ruminococcus_1 0.41204 0.02279 18.07756 0.00000
intB:time Ruminococcus_1 -0.32122 0.03226 -9.95826 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.8807
Significance test not available for random effectsgenus.fit5b <- glmm_microbiome(mydata=microbiome_data,model.number=5,
taxa.level="Genus", taxa.subset = backNames,
link="negbinom",
model="1 + intB + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 1.976585e-11
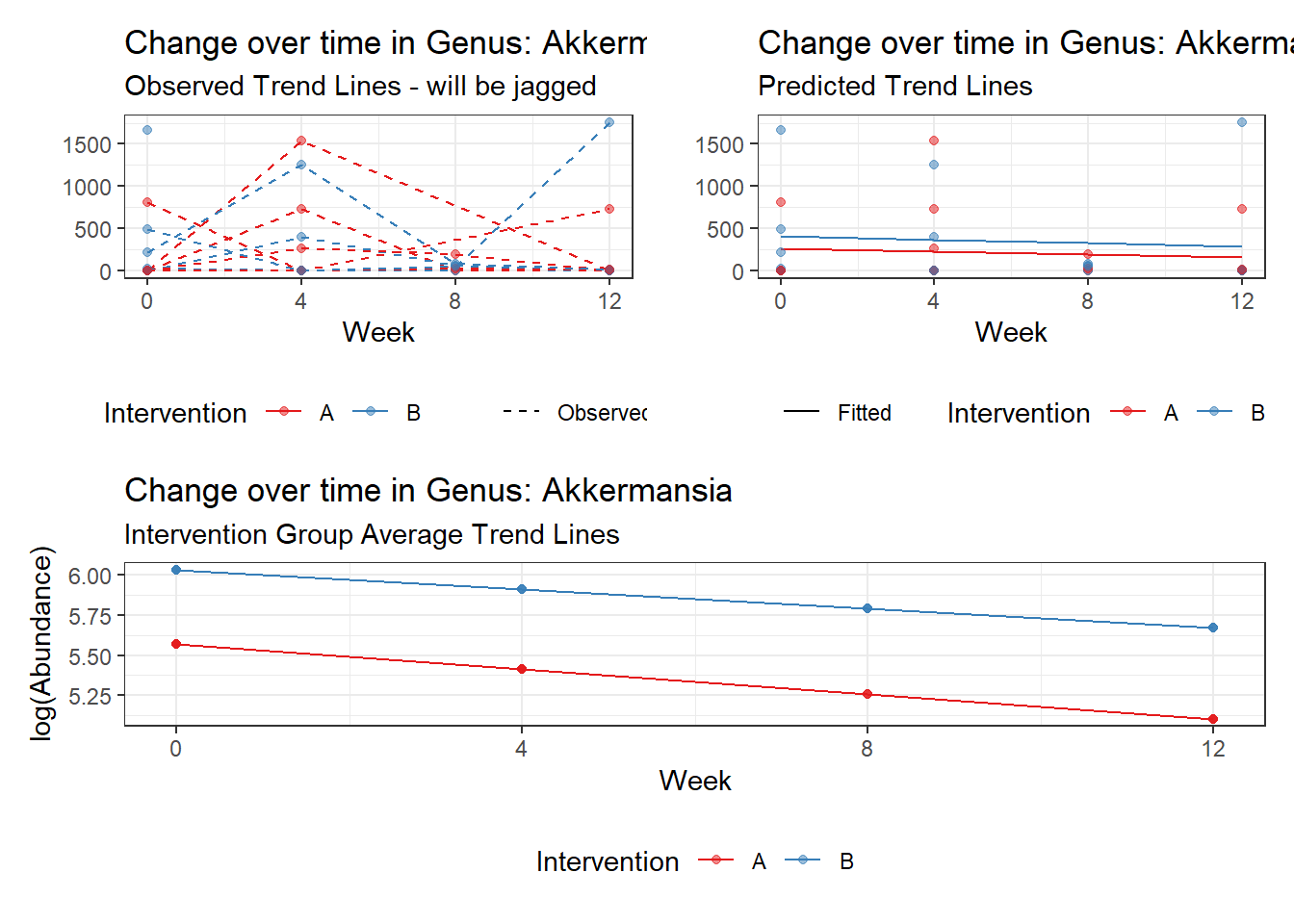
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 5.56968 0.96093 5.79616 0.00000
intB Akkermansia 0.45954 1.33004 0.34551 0.72971
time Akkermansia -0.15489 0.57324 -0.27020 0.78701
intB:time Akkermansia 0.03610 0.74911 0.04819 0.96157
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.4459e-06
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinom
Intraclass Correlation (ICC): 0.2210869

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.86278 0.23993 32.77095 0.00000
intB Bacteroides -0.36350 0.35881 -1.01307 0.31103
time Bacteroides -0.04319 0.05877 -0.73491 0.46240
intB:time Bacteroides -0.04849 0.08926 -0.54327 0.58695
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.53277
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinom
Intraclass Correlation (ICC): 0.2649347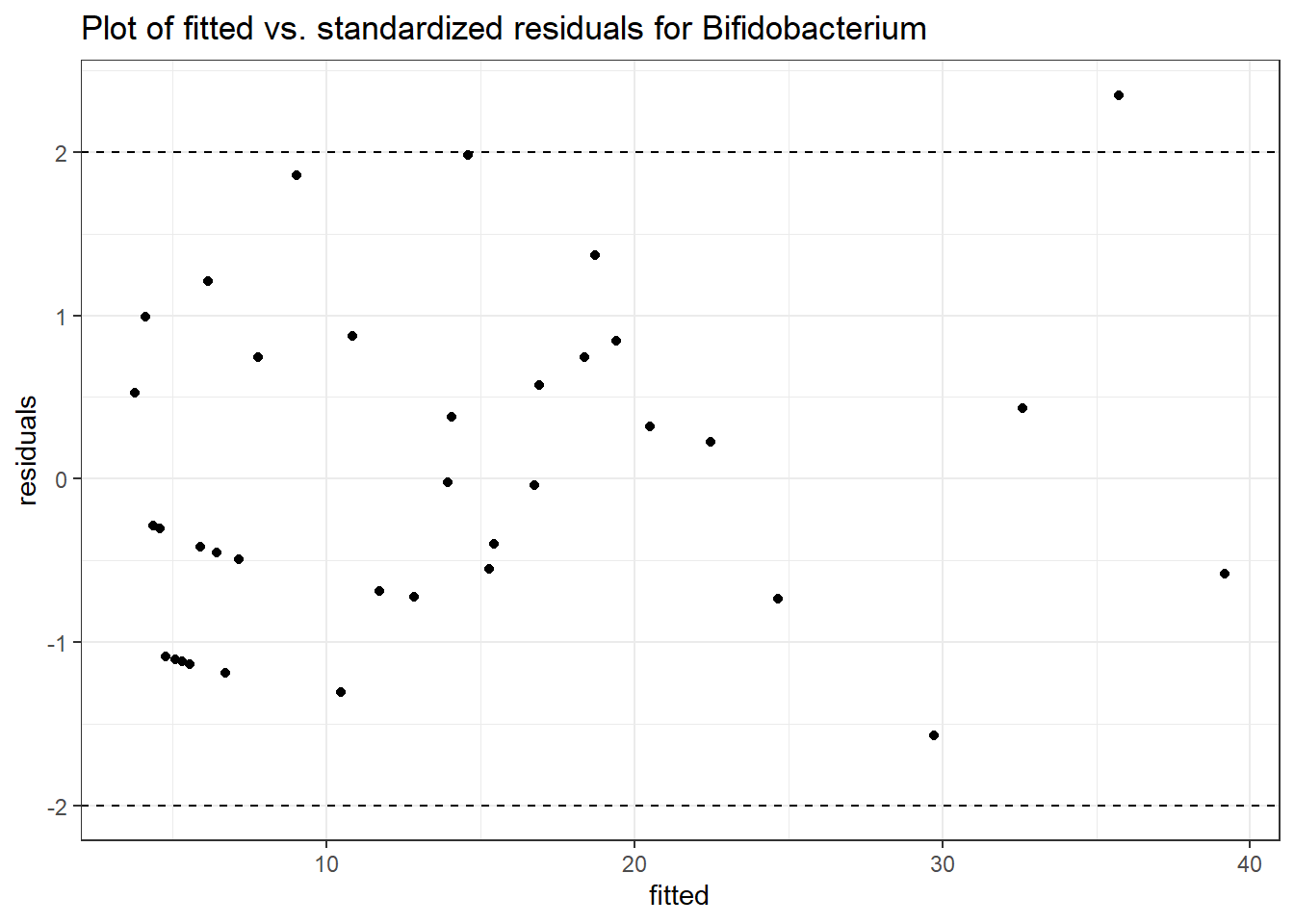
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.13534 0.73198 2.91720 0.00353
intB Bifidobacterium 0.74018 1.00563 0.73604 0.46171
time Bifidobacterium -0.14834 0.37423 -0.39640 0.69181
intB:time Bifidobacterium 0.24025 0.55370 0.43390 0.66436
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.60035
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + intB + time + intB:time + (1 |
SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomboundary (singular) fit: see ?isSingular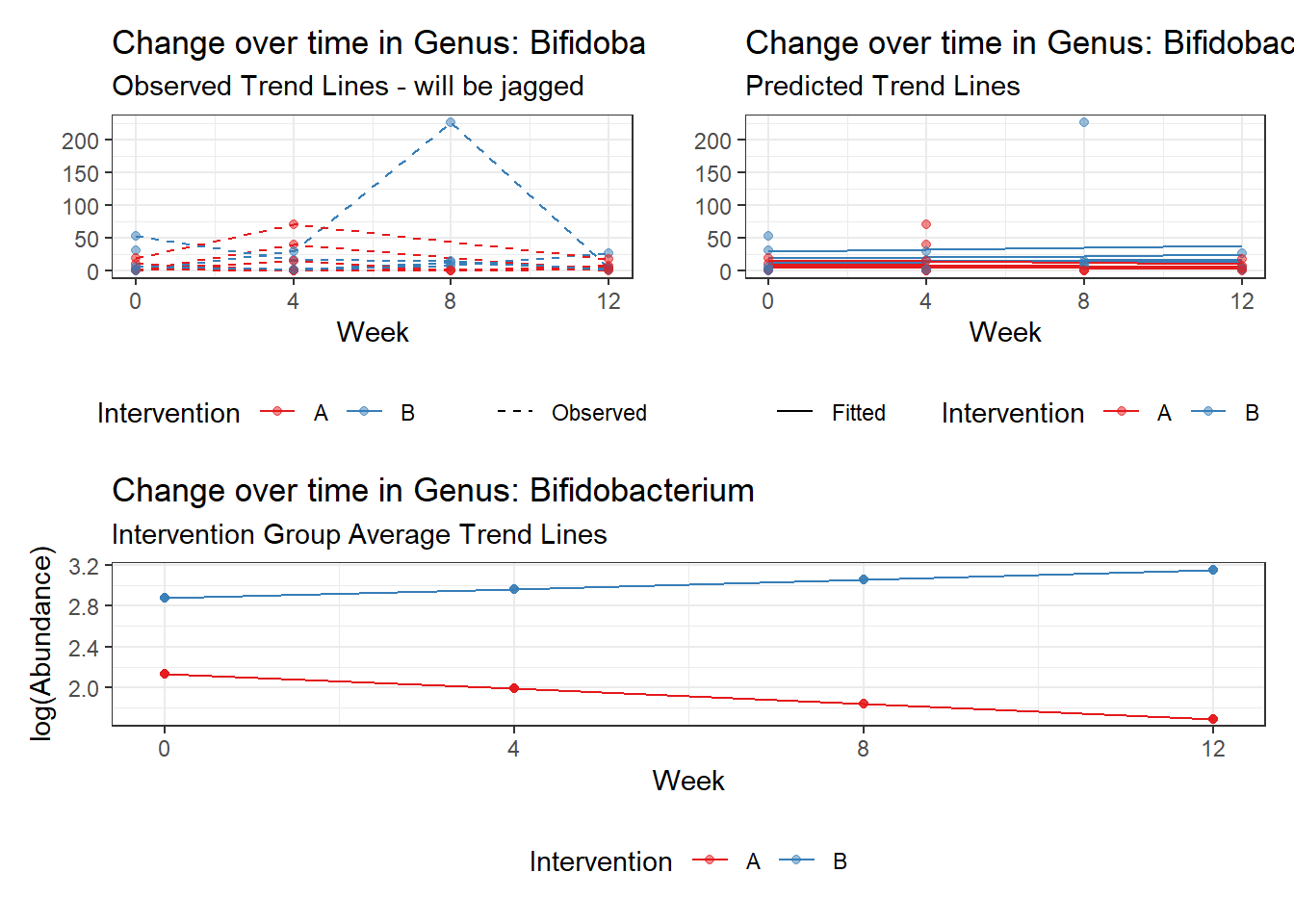
Intraclass Correlation (ICC): 8.491197e-11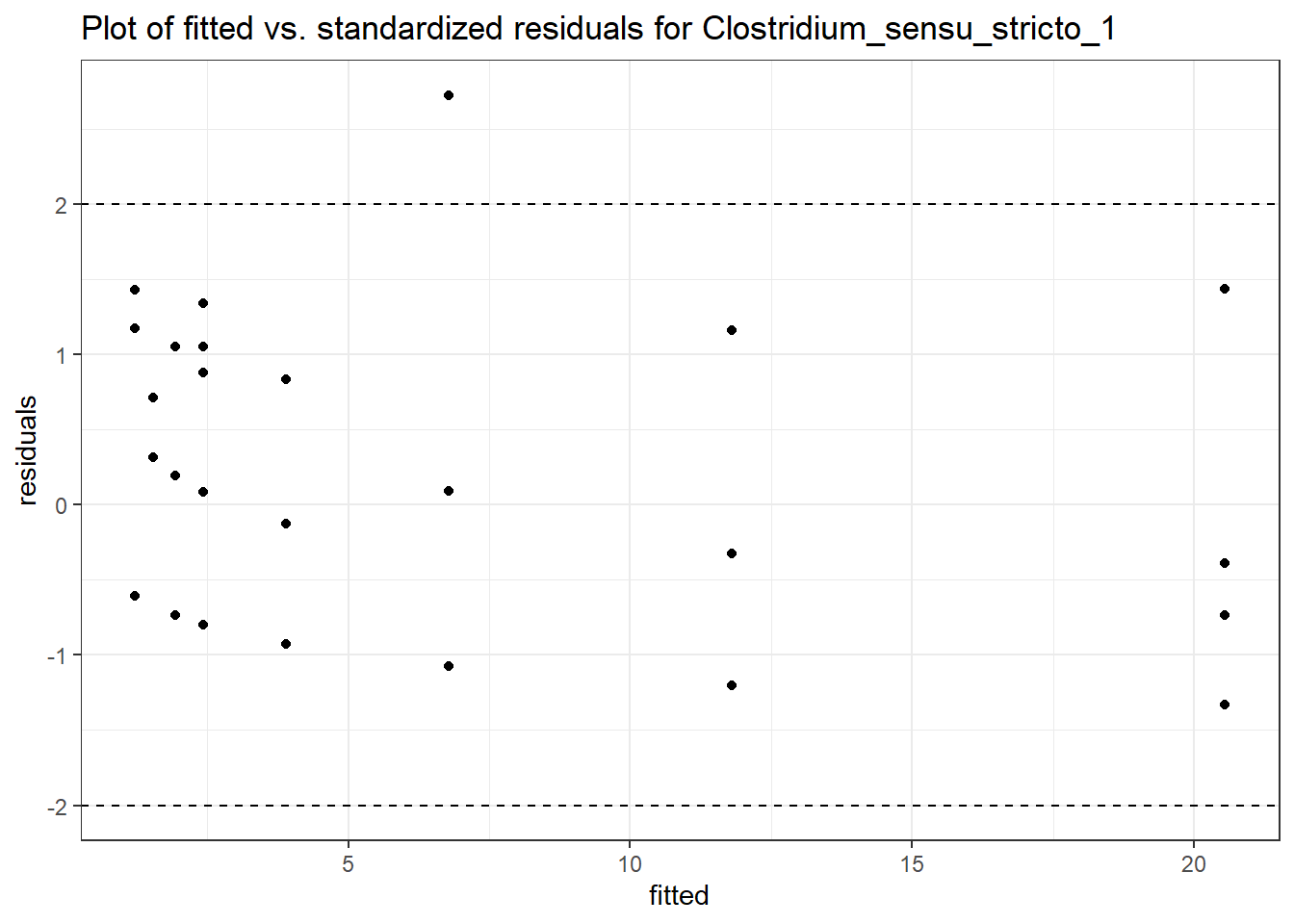
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.88902 0.63128 1.40828 0.15905
intB Clostridium_sensu_stricto_1 0.47097 1.01863 0.46235 0.64383
time Clostridium_sensu_stricto_1 -0.23107 0.35662 -0.64795 0.51701
intB:time Clostridium_sensu_stricto_1 0.78538 0.58882 1.33383 0.18226
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 9.2148e-06
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 8.833692e-13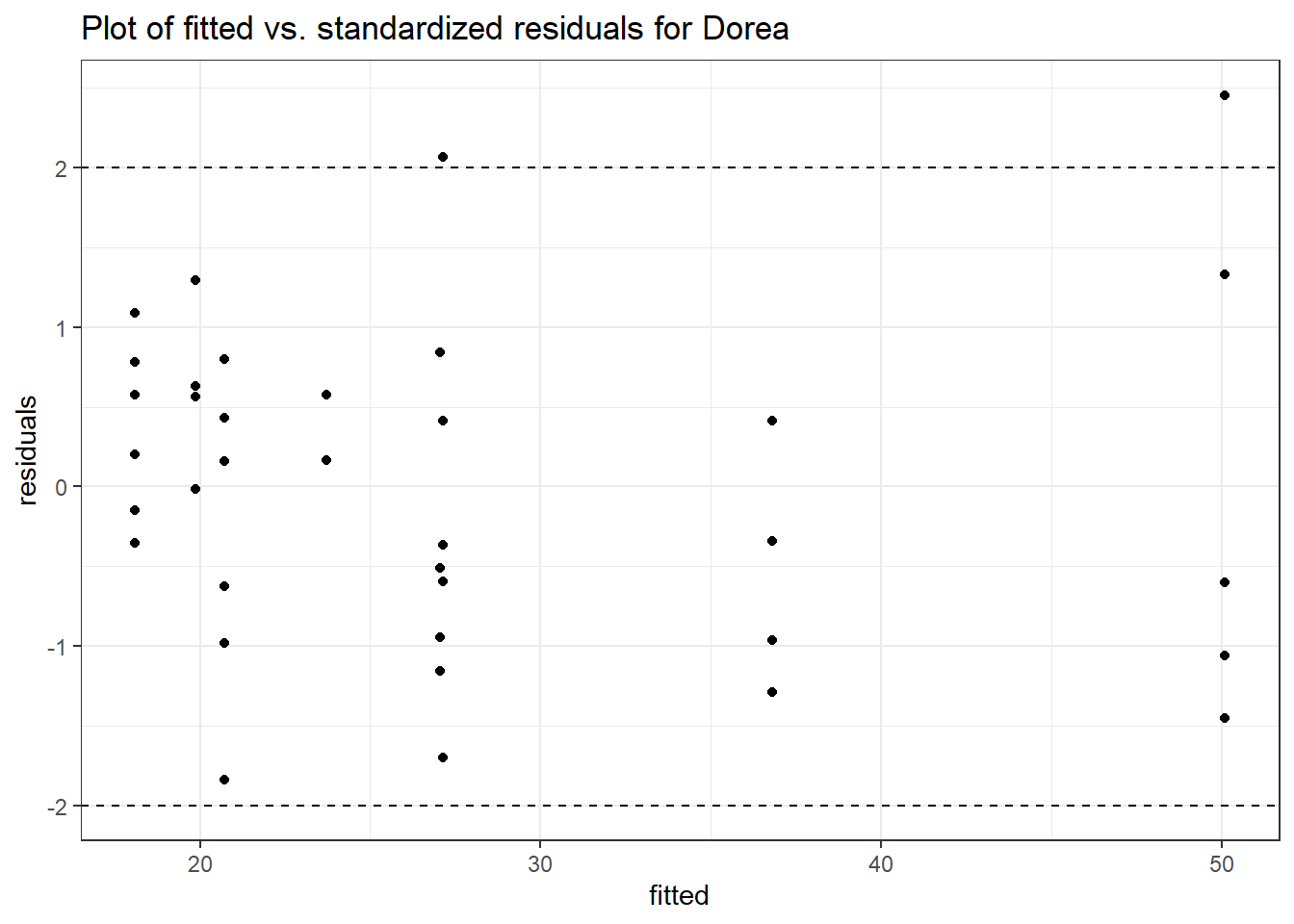
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 2.89556 0.22561 12.83439 0.00000
intB Dorea 1.01860 0.32568 3.12763 0.00176
time Dorea 0.13505 0.12398 1.08936 0.27600
intB:time Dorea -0.44344 0.17680 -2.50813 0.01214
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 9.3988e-07
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomboundary (singular) fit: see ?isSingular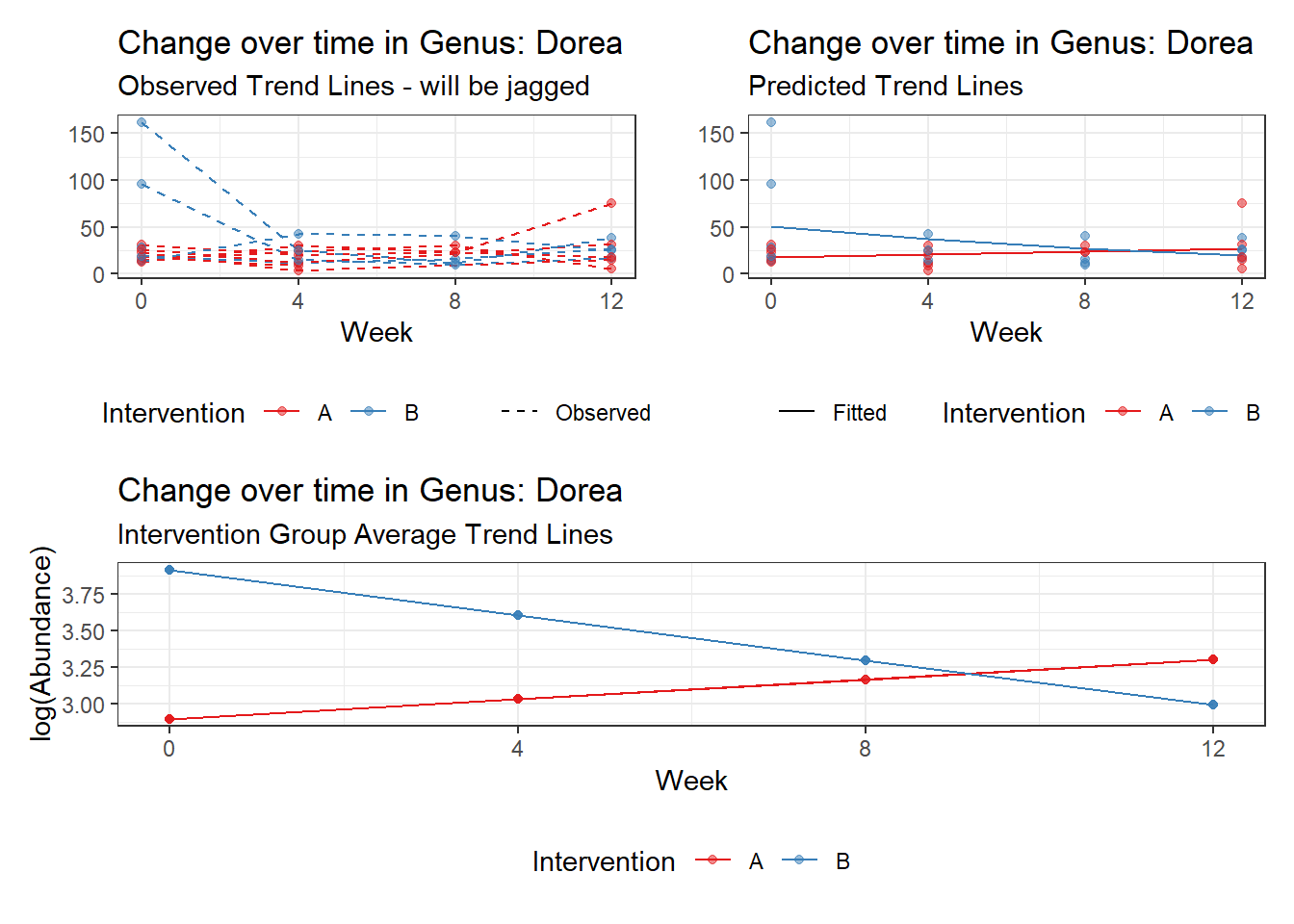
Intraclass Correlation (ICC): 1.843875e-13
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.72260 0.35413 18.98333 0.00000
intB Faecalibacterium -0.06372 0.52760 -0.12078 0.90387
time Faecalibacterium 0.02226 0.20926 0.10639 0.91528
intB:time Faecalibacterium -0.14079 0.30598 -0.46013 0.64543
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.294e-07
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced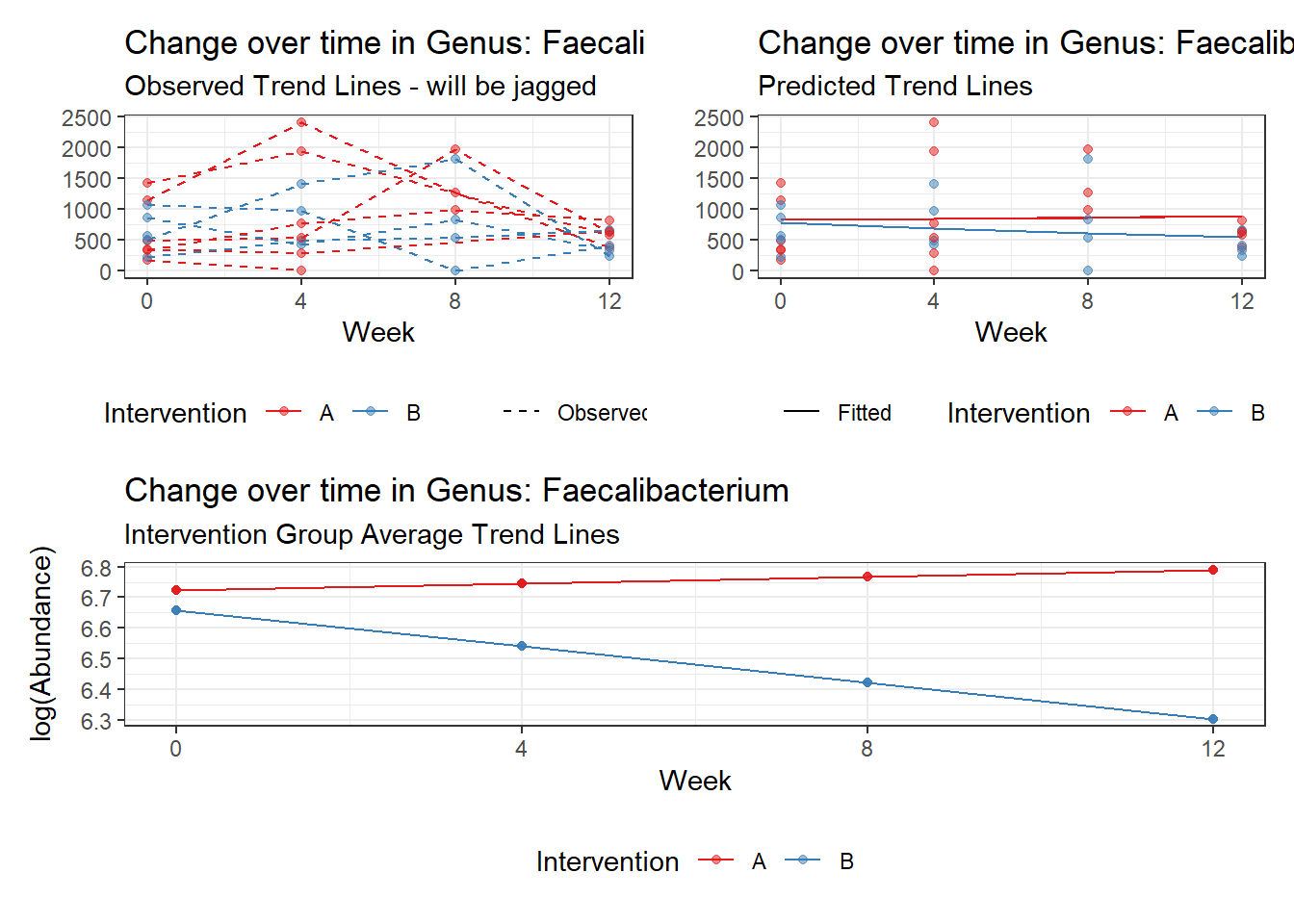
Intraclass Correlation (ICC): 0.2828738
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.21533 0.25977 16.22735 0.00000
intB Lachnospira -0.96605 0.38989 -2.47778 0.01322
time Lachnospira 0.07061 0.02214 3.18901 0.00143
intB:time Lachnospira 0.13991 0.03753 3.72811 0.00019
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.62806
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.011536 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular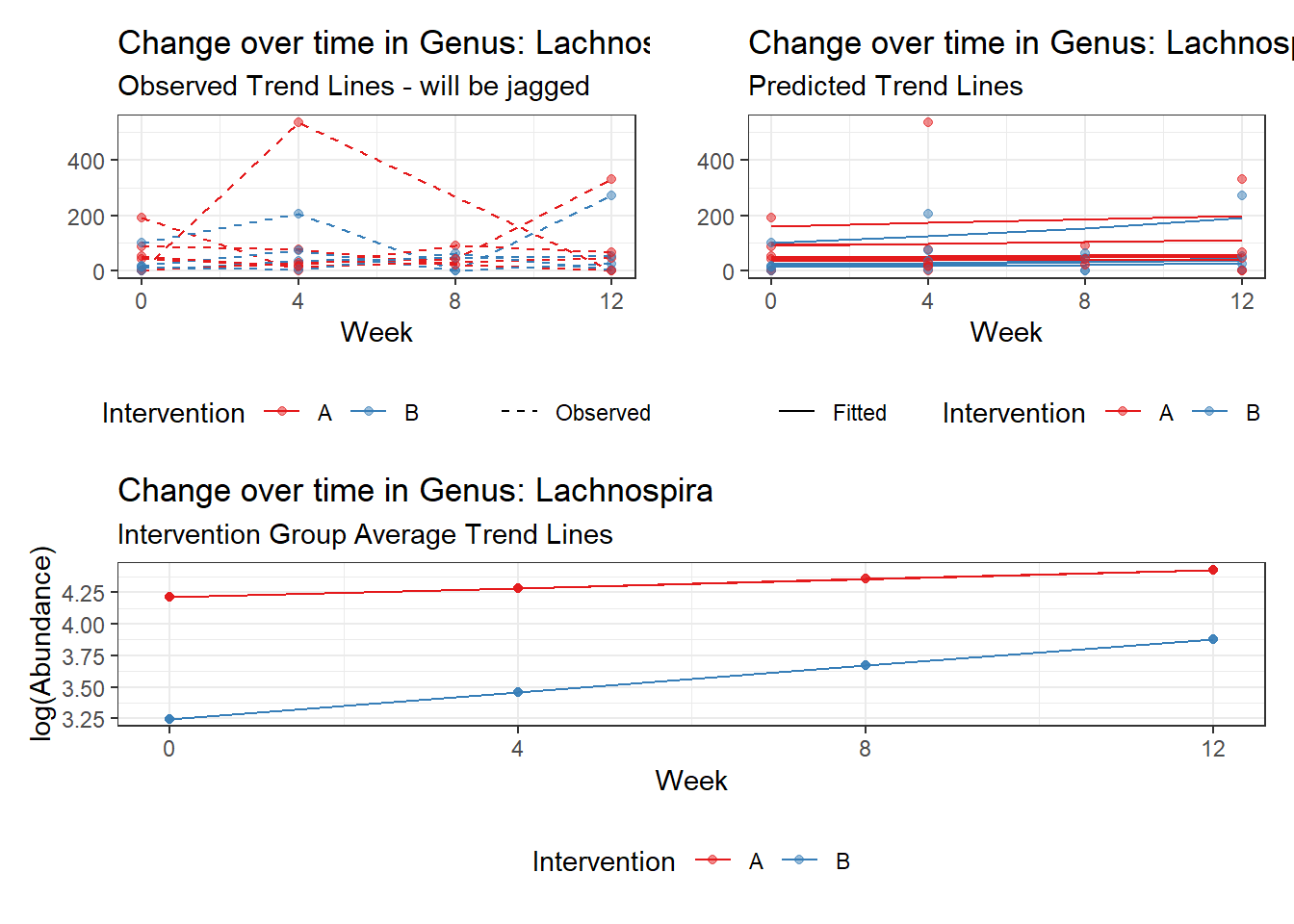
Intraclass Correlation (ICC): 7.843828e-12
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus 3.56743 1.36352 2.61633 0.00889
intB Lactobacillus -5.19627 2.20261 -2.35915 0.01832
time Lactobacillus -1.68879 0.89044 -1.89657 0.05788
intB:time Lactobacillus 1.61174 1.34608 1.19736 0.23117
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.8007e-06
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00766701 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 5.254418e-13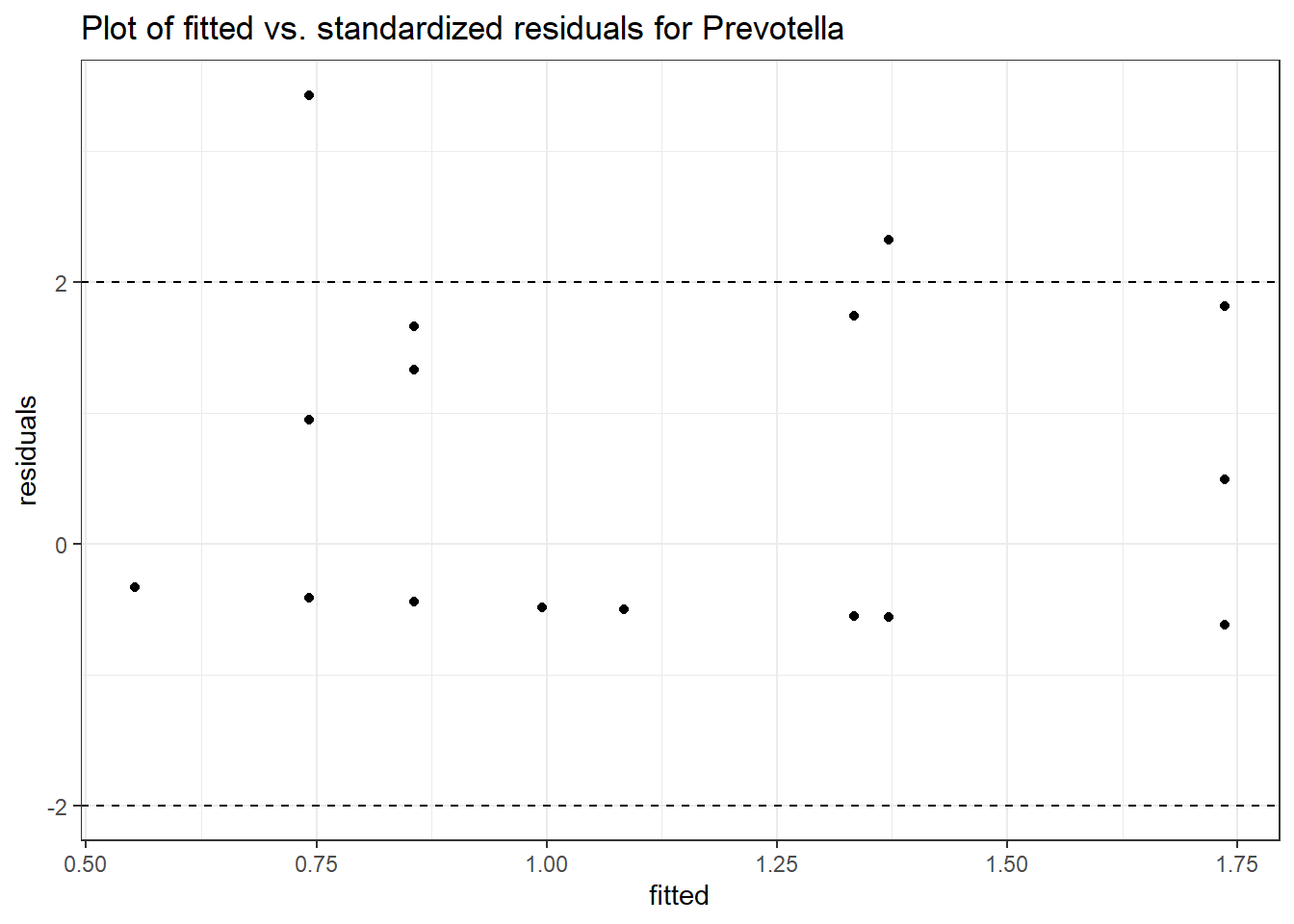
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -0.59218 1.37499 -0.43068 0.66670
intB Prevotella 1.14402 1.81176 0.63144 0.52775
time Prevotella 0.29344 0.85101 0.34482 0.73023
intB:time Prevotella -0.52914 1.06756 -0.49565 0.62014
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 7.2487e-07
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomboundary (singular) fit: see ?isSingular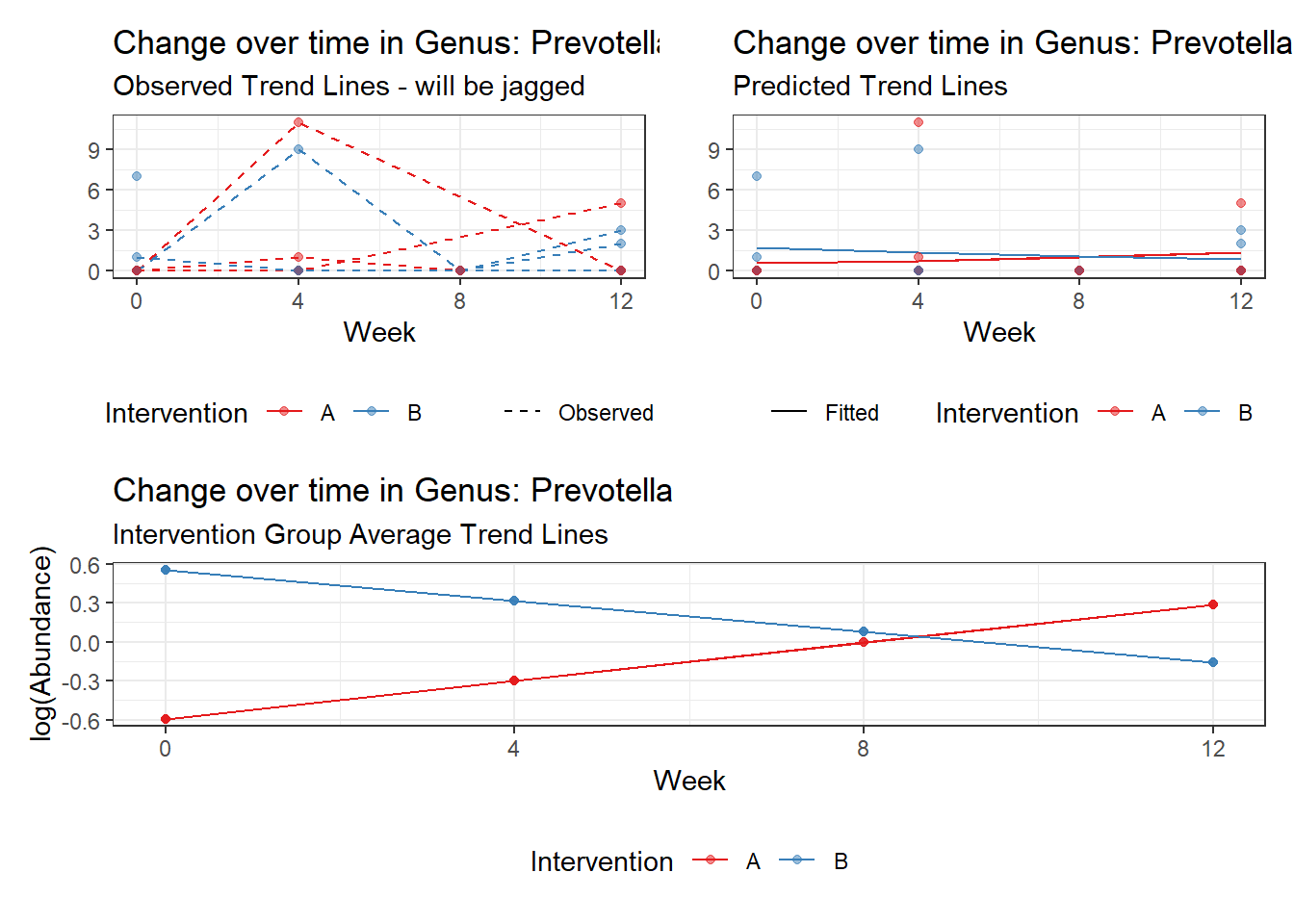
Intraclass Correlation (ICC): 1.915704e-11
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 2.96787 0.50931 5.82723 0.00000
intB Roseburia -0.05790 0.75481 -0.07671 0.93885
time Roseburia 0.13106 0.28262 0.46371 0.64285
intB:time Roseburia 0.01222 0.41021 0.02979 0.97624
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.3769e-06
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000352d7578>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0192163 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular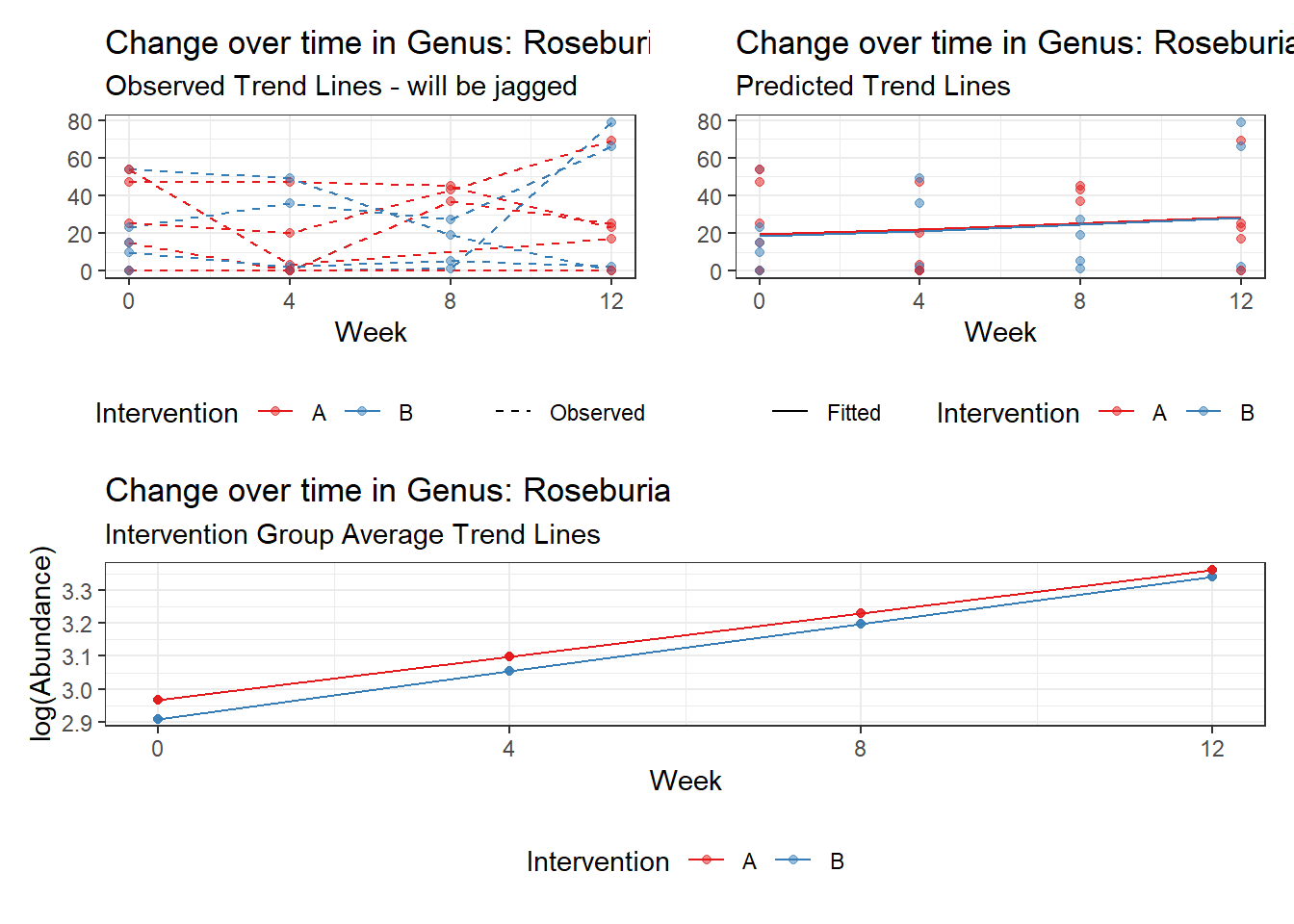
Intraclass Correlation (ICC): 1.105465e-11
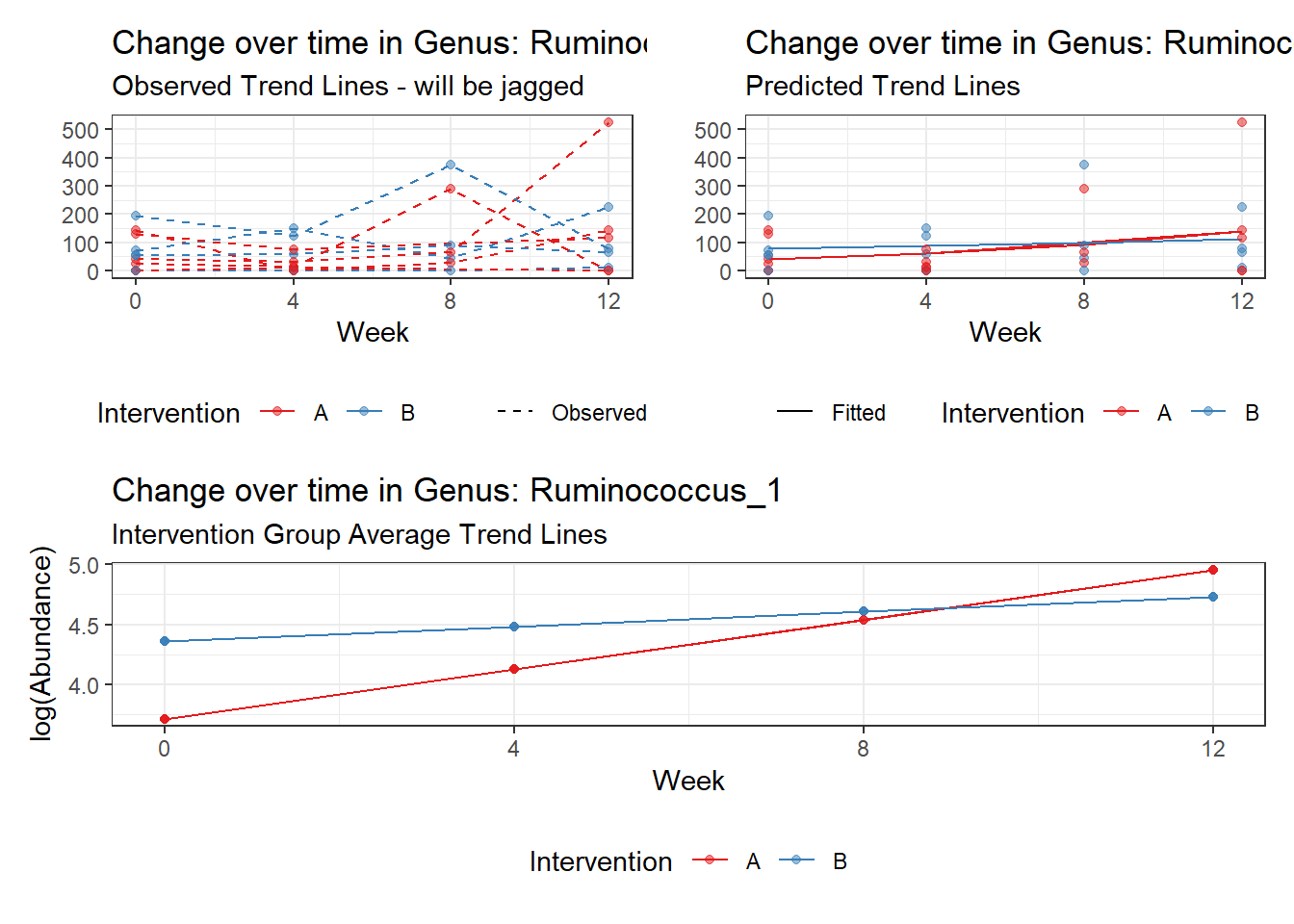
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 3.71192 0.54351 6.82957 0.00000
intB Ruminococcus_1 0.64950 0.85210 0.76223 0.44592
time Ruminococcus_1 0.41404 0.29315 1.41238 0.15784
intB:time Ruminococcus_1 -0.29160 0.46969 -0.62083 0.53471
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.3249e-06
Significance test not available for random effectsCovariate(s): Time by intervention interaction (only)
Only the interaction term.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
genus.fit6 <- glmm_microbiome(mydata=microbiome_data,model.number=6,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.8603956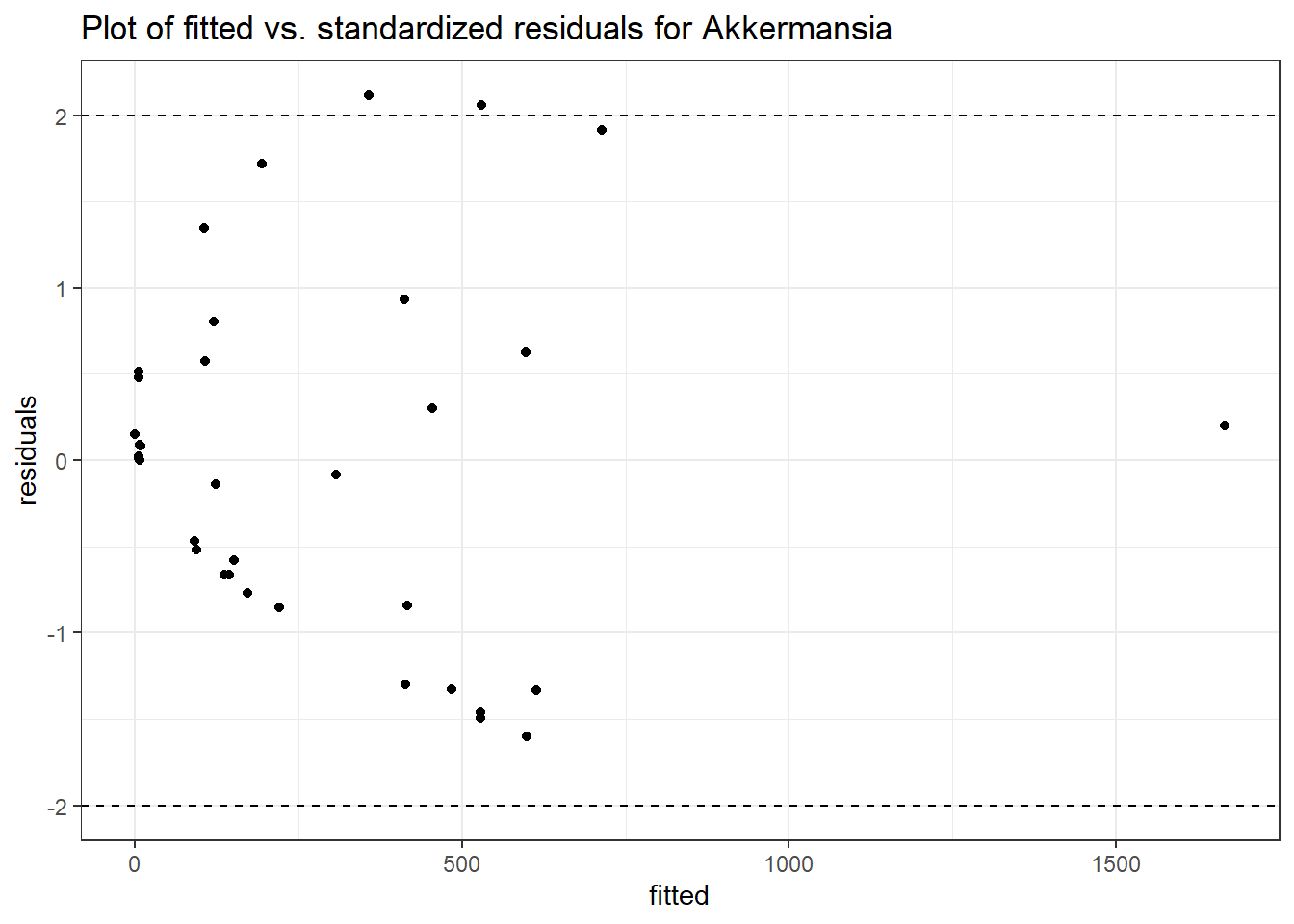

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 4.49776 0.75515 5.95610 0
time Akkermansia -0.12318 0.01296 -9.50252 0
time:intB Akkermansia 0.27351 0.01882 14.52988 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4826
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.2806381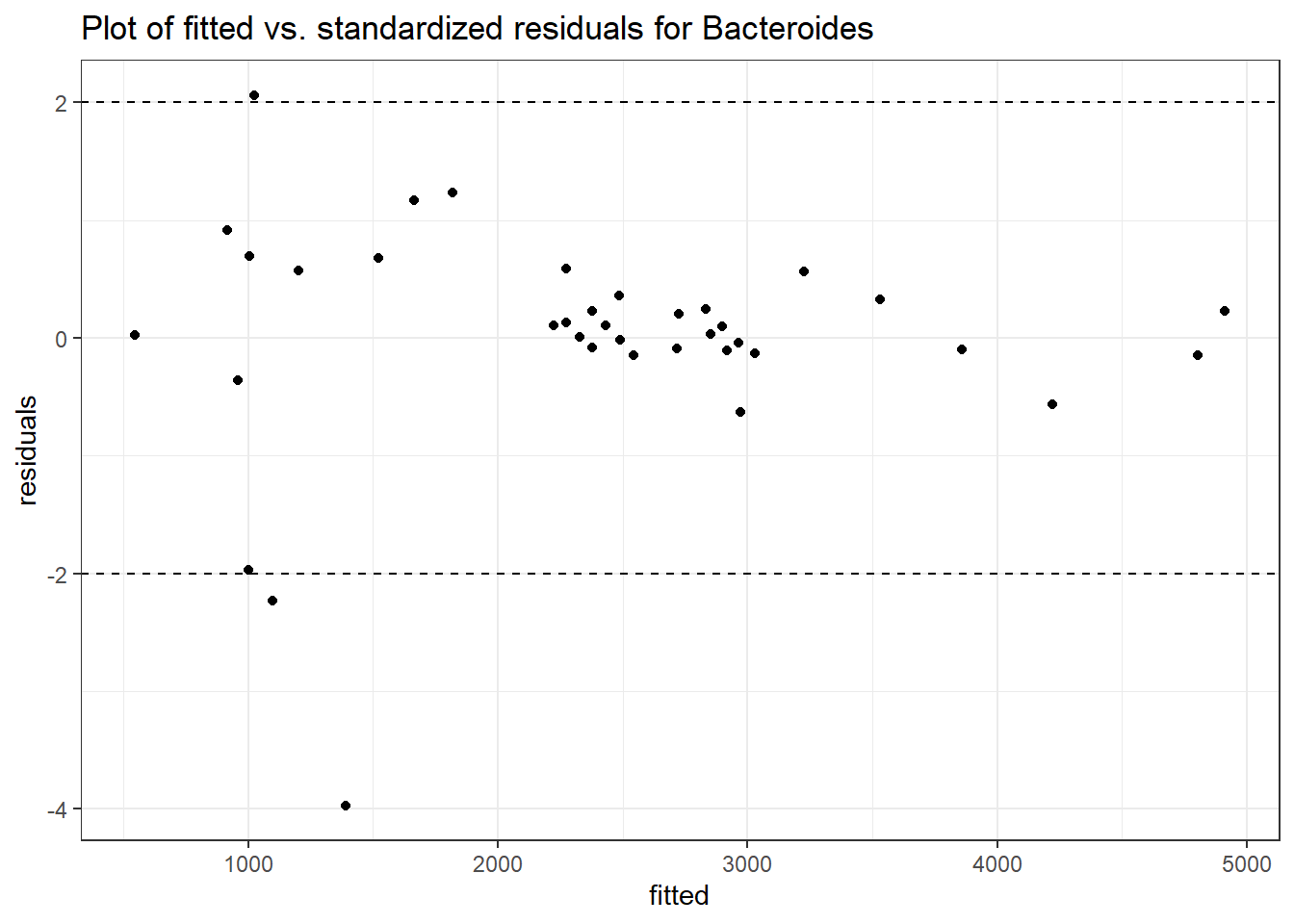
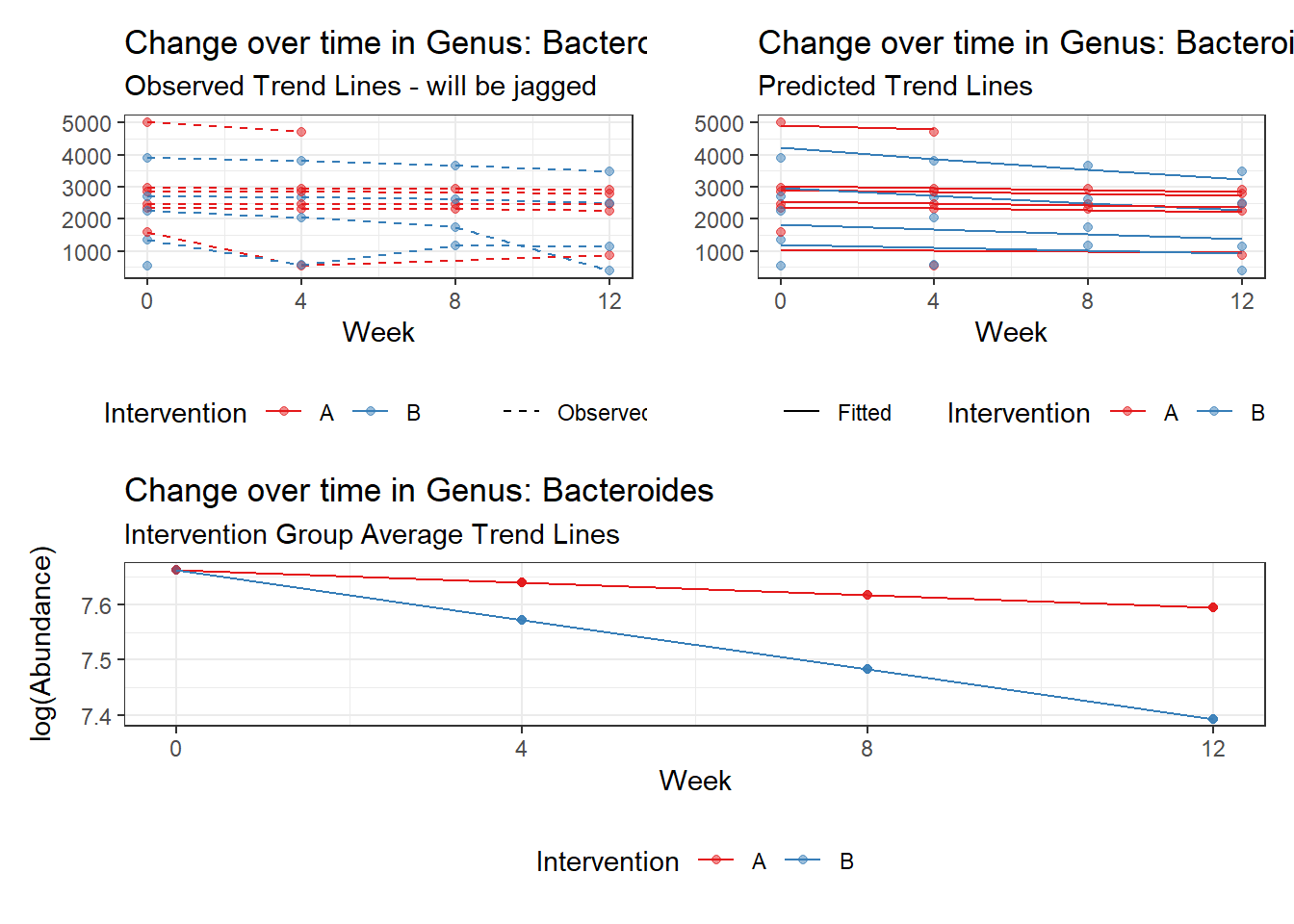
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.66227 0.18843 40.66363 0
time Bacteroides -0.02253 0.00413 -5.45070 0
time:intB Bacteroides -0.06706 0.00629 -10.66860 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.6246
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.5384285

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.28210 0.33594 6.79313 0.00000
time Bifidobacterium -0.12149 0.06172 -1.96853 0.04901
time:intB Bifidobacterium 0.26303 0.07578 3.47096 0.00052
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0801
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.6120341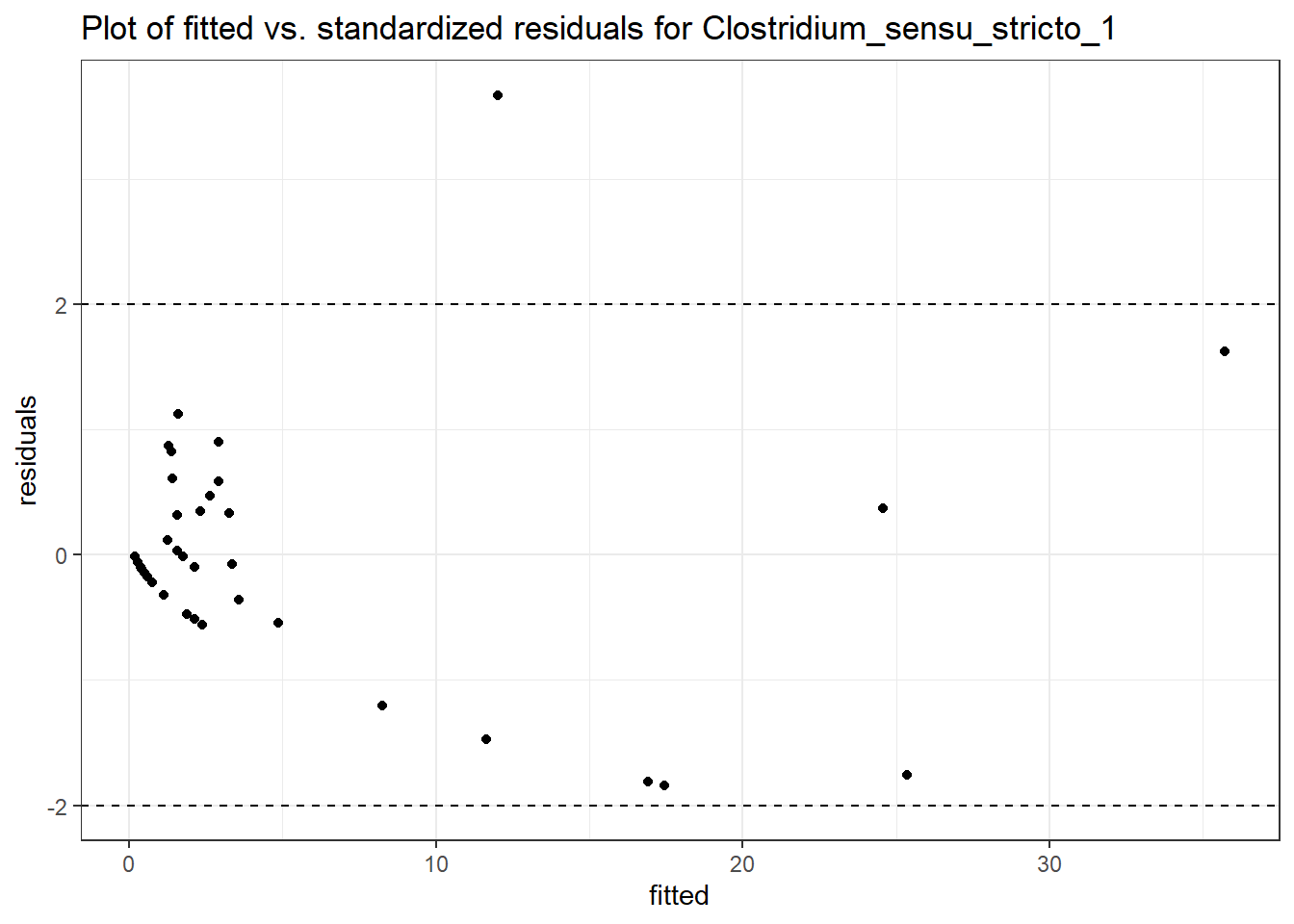
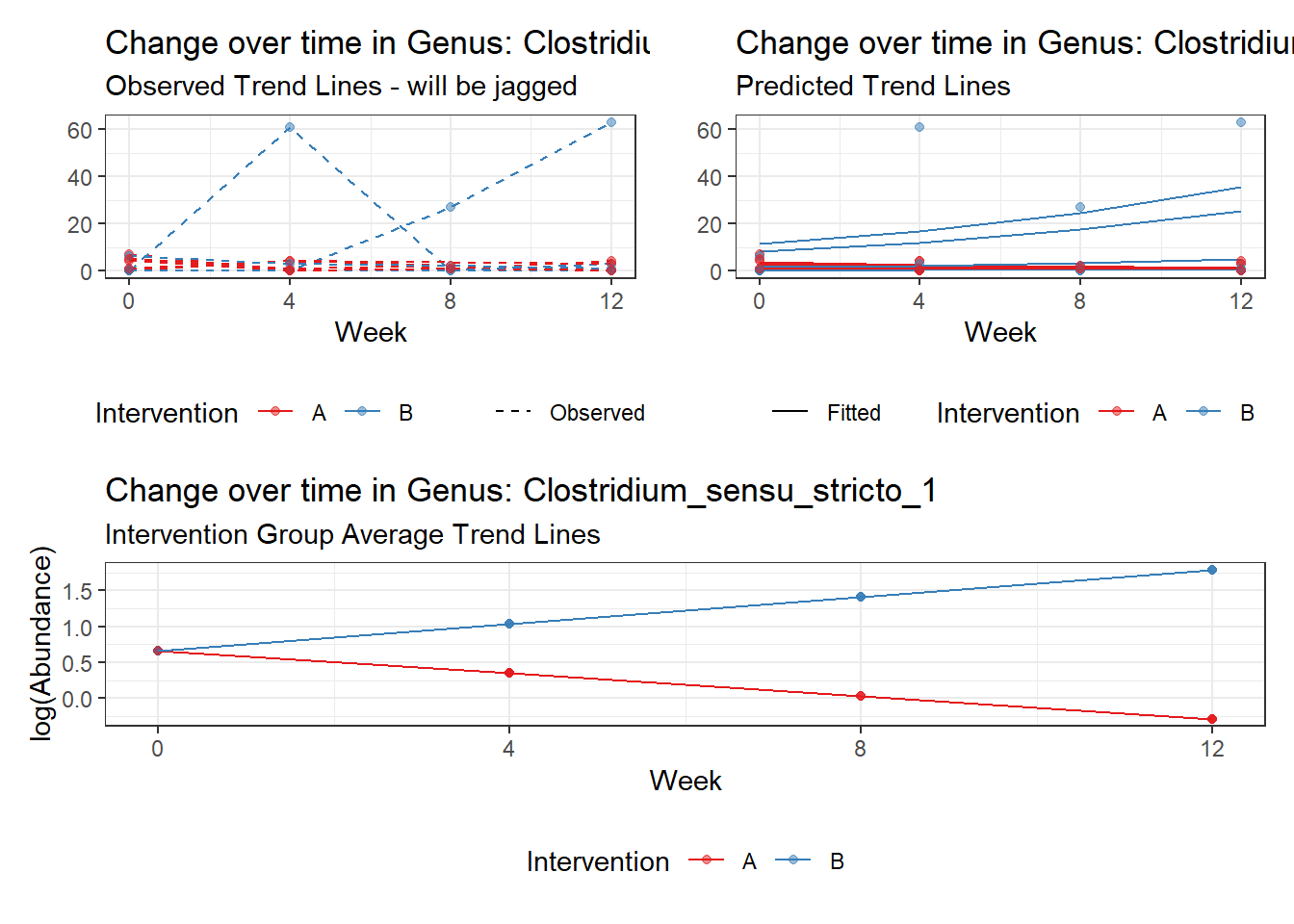
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.66218 0.42862 1.54490 0.12237
time Clostridium_sensu_stricto_1 -0.31475 0.14725 -2.13746 0.03256
time:intB Clostridium_sensu_stricto_1 0.68849 0.16235 4.24083 0.00002
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.256
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.2833984
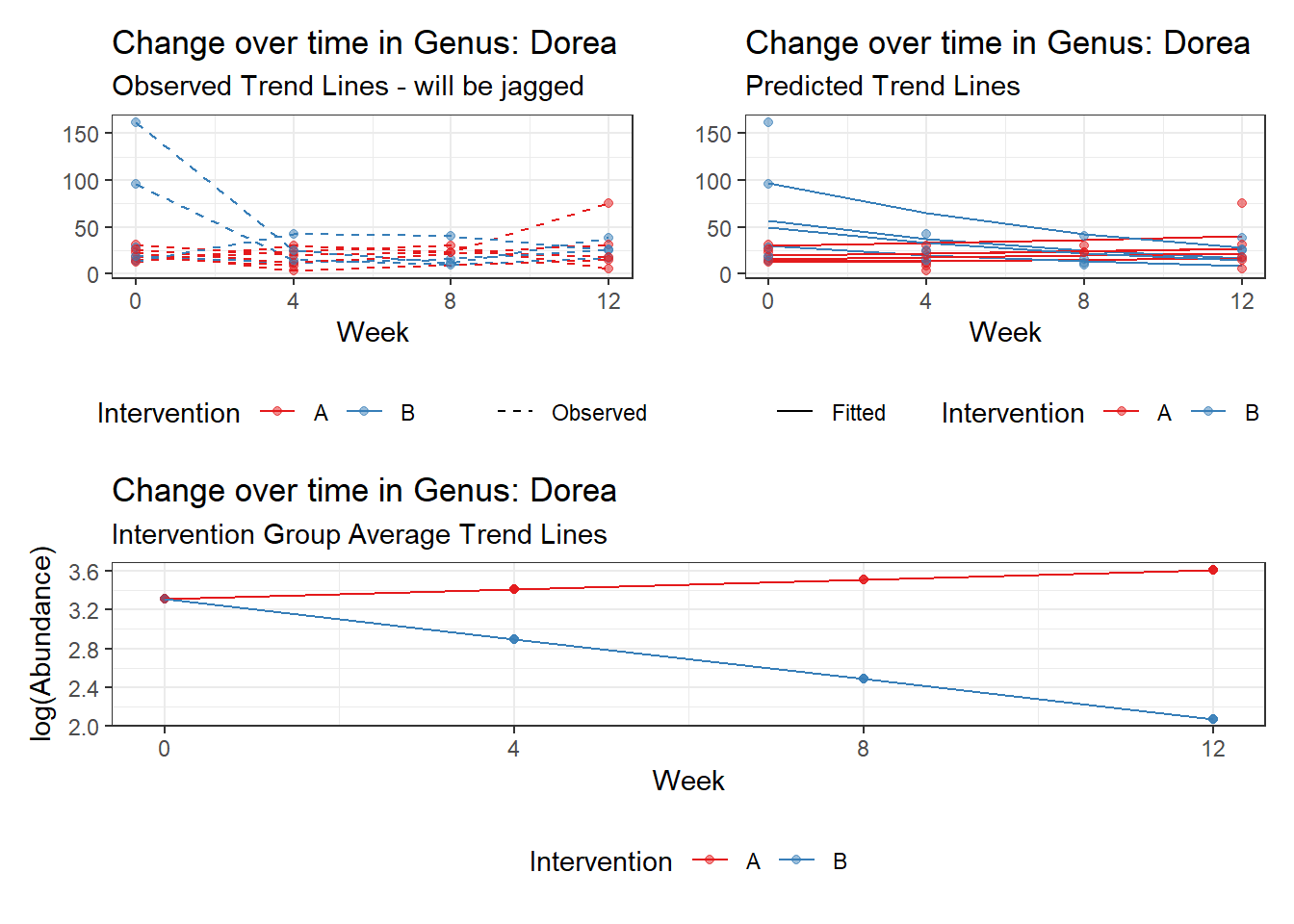
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.31091 0.19662 16.83886 0.00000
time Dorea 0.09951 0.04178 2.38165 0.01724
time:intB Dorea -0.51134 0.05839 -8.75697 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.62887
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.3438831
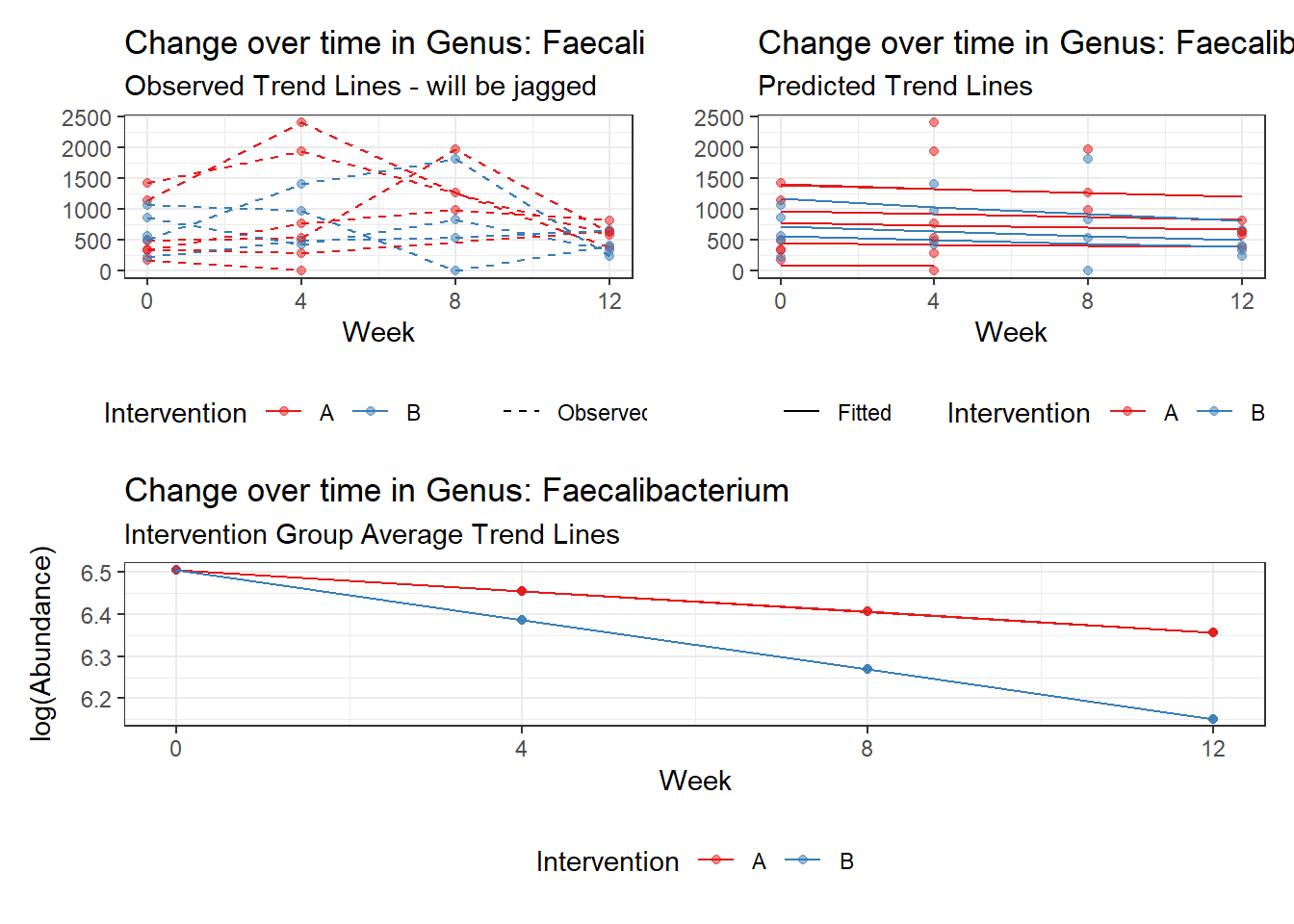
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.50537 0.21857 29.76312 0
time Faecalibacterium -0.04938 0.00665 -7.42311 0
time:intB Faecalibacterium -0.06861 0.01094 -6.27395 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.72396
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.3816131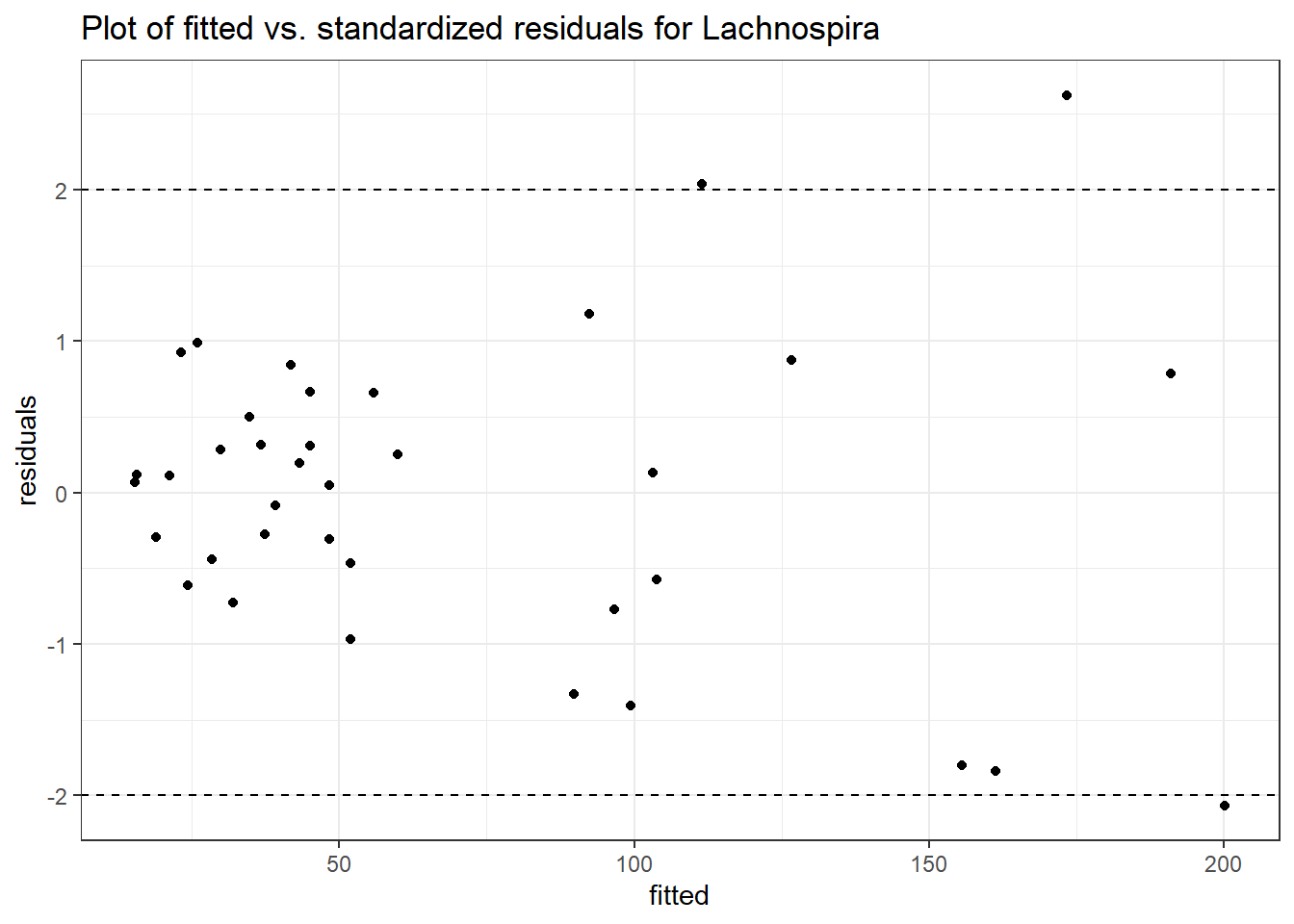

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 3.77971 0.24027 15.73100 0.00000
time Lachnospira 0.07211 0.02183 3.30276 0.00096
time:intB Lachnospira 0.13308 0.03699 3.59745 0.00032
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.78556
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.8858612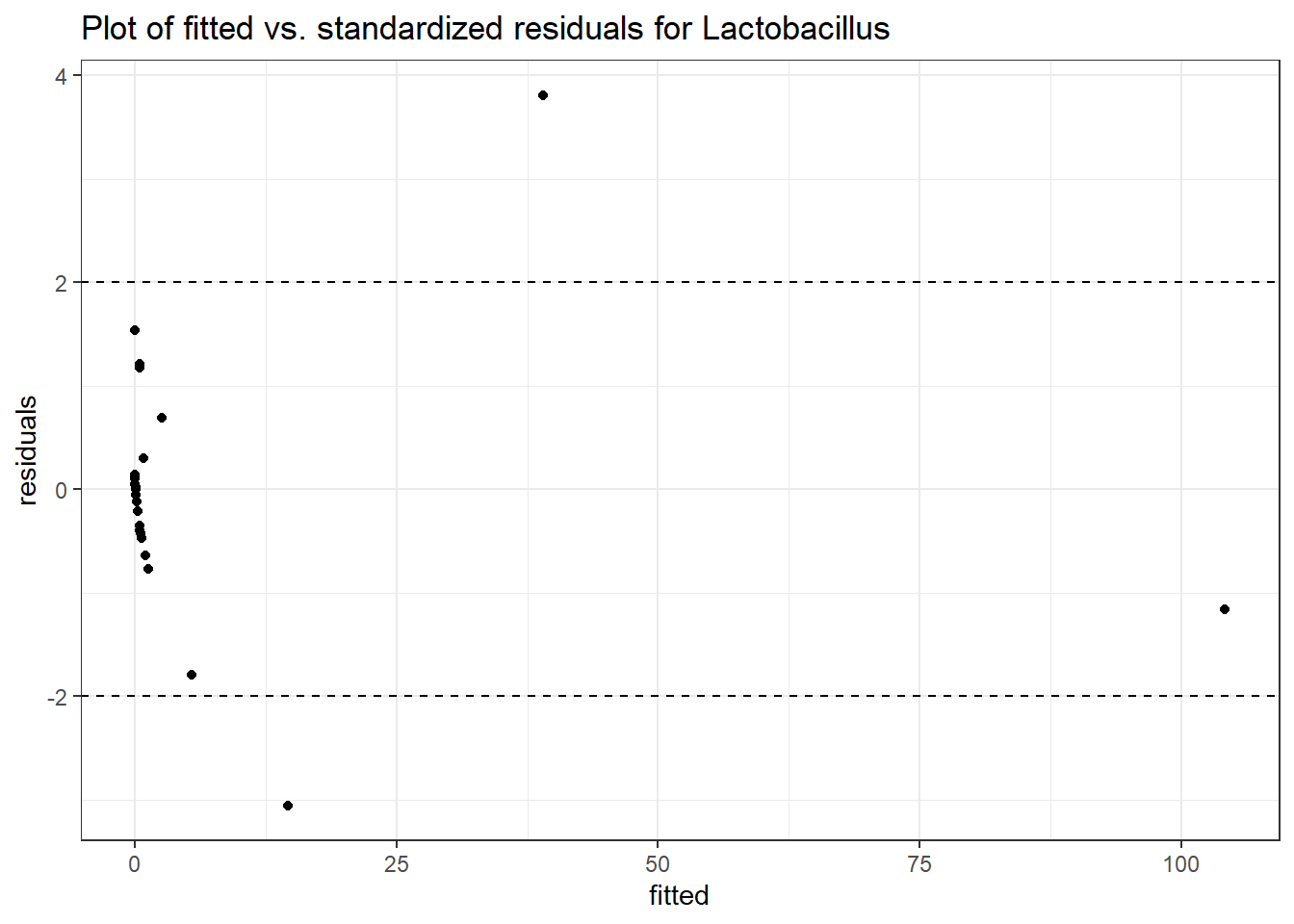

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus -1.37721 1.19229 -1.15509 0.24805
time Lactobacillus -0.98338 0.09738 -10.09861 0.00000
time:intB Lactobacillus 0.89604 0.52768 1.69809 0.08949
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.7859
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.7675638
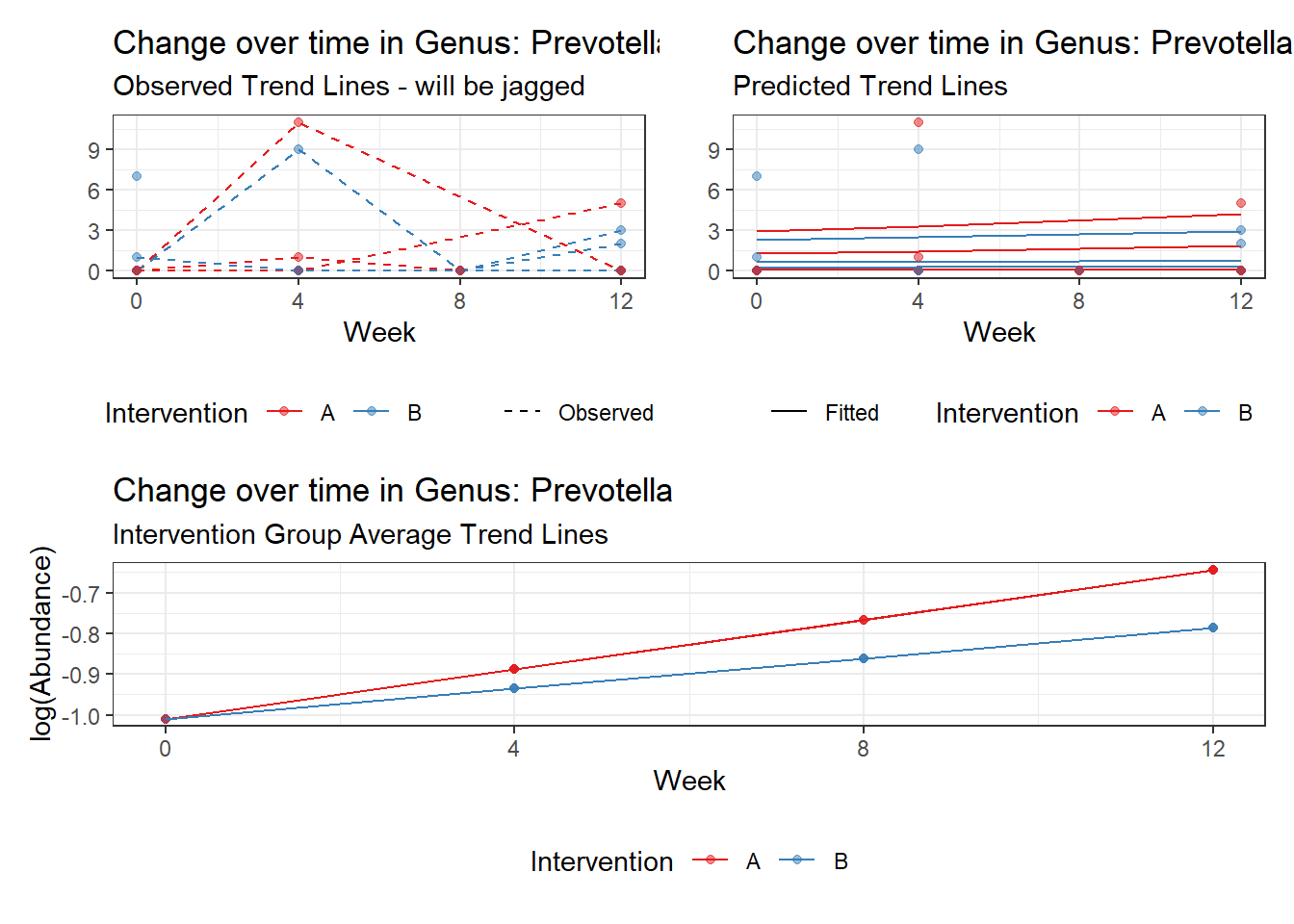
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.00942 0.73528 -1.37283 0.16980
time Prevotella 0.12169 0.18724 0.64995 0.51572
time:intB Prevotella -0.04722 0.28353 -0.16655 0.86772
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.8172
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.7535659
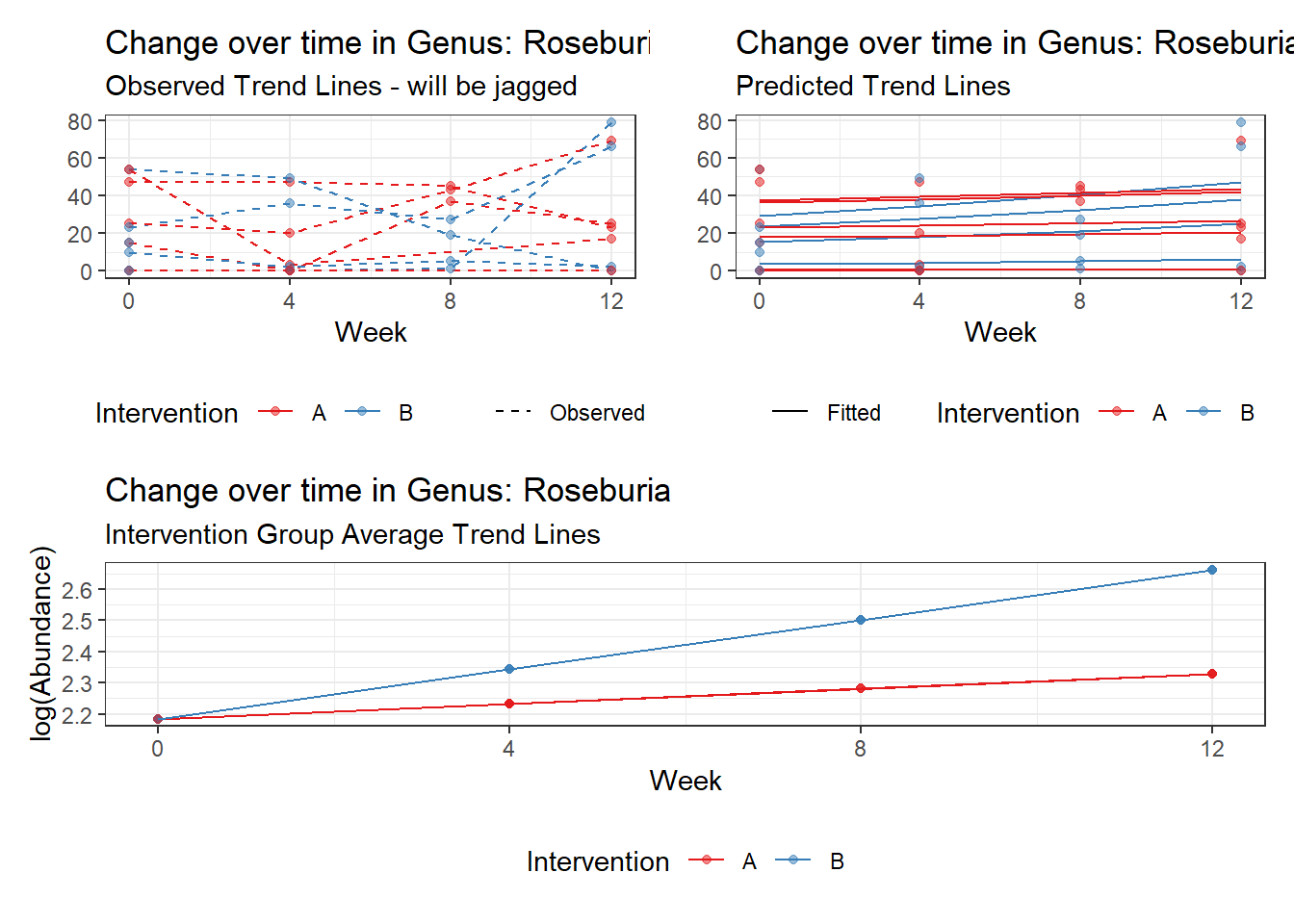
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 2.18489 0.54845 3.98373 0.00007
time Roseburia 0.04811 0.04027 1.19459 0.23225
time:intB Roseburia 0.11068 0.06150 1.79955 0.07193
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7487
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x00000000055459f8>
Link: poisson
Intraclass Correlation (ICC): 0.7953052
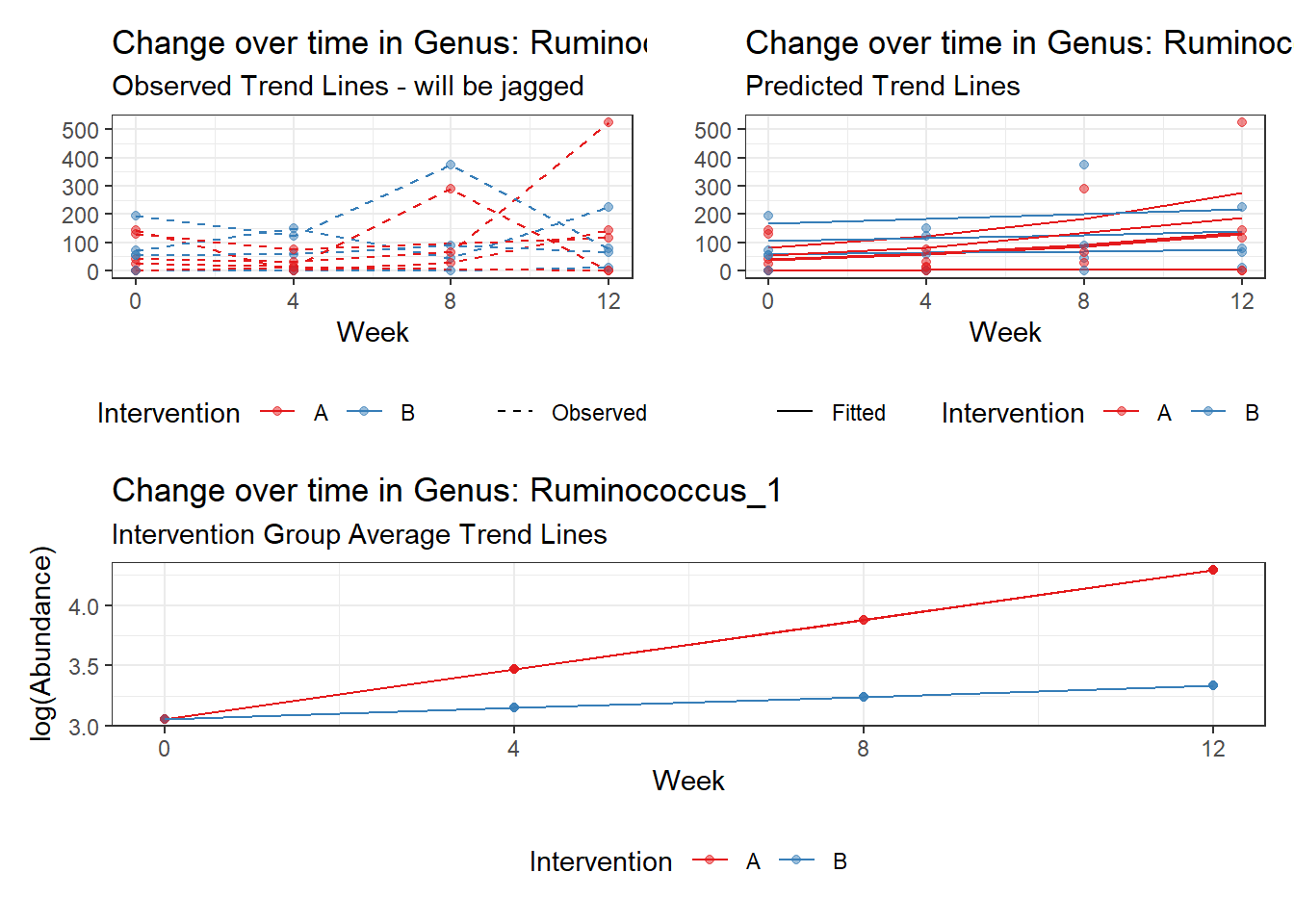
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 3.05936 0.60321 5.07180 0
time Ruminococcus_1 0.41119 0.02277 18.06152 0
time:intB Ruminococcus_1 -0.31978 0.03222 -9.92361 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.9711
Significance test not available for random effectsgenus.fit6b <- glmm_microbiome(mydata=microbiome_data,model.number=6,
taxa.level="Genus", taxa.subset = backNames,
link="negbinom",
model="1 + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 8.057189e-12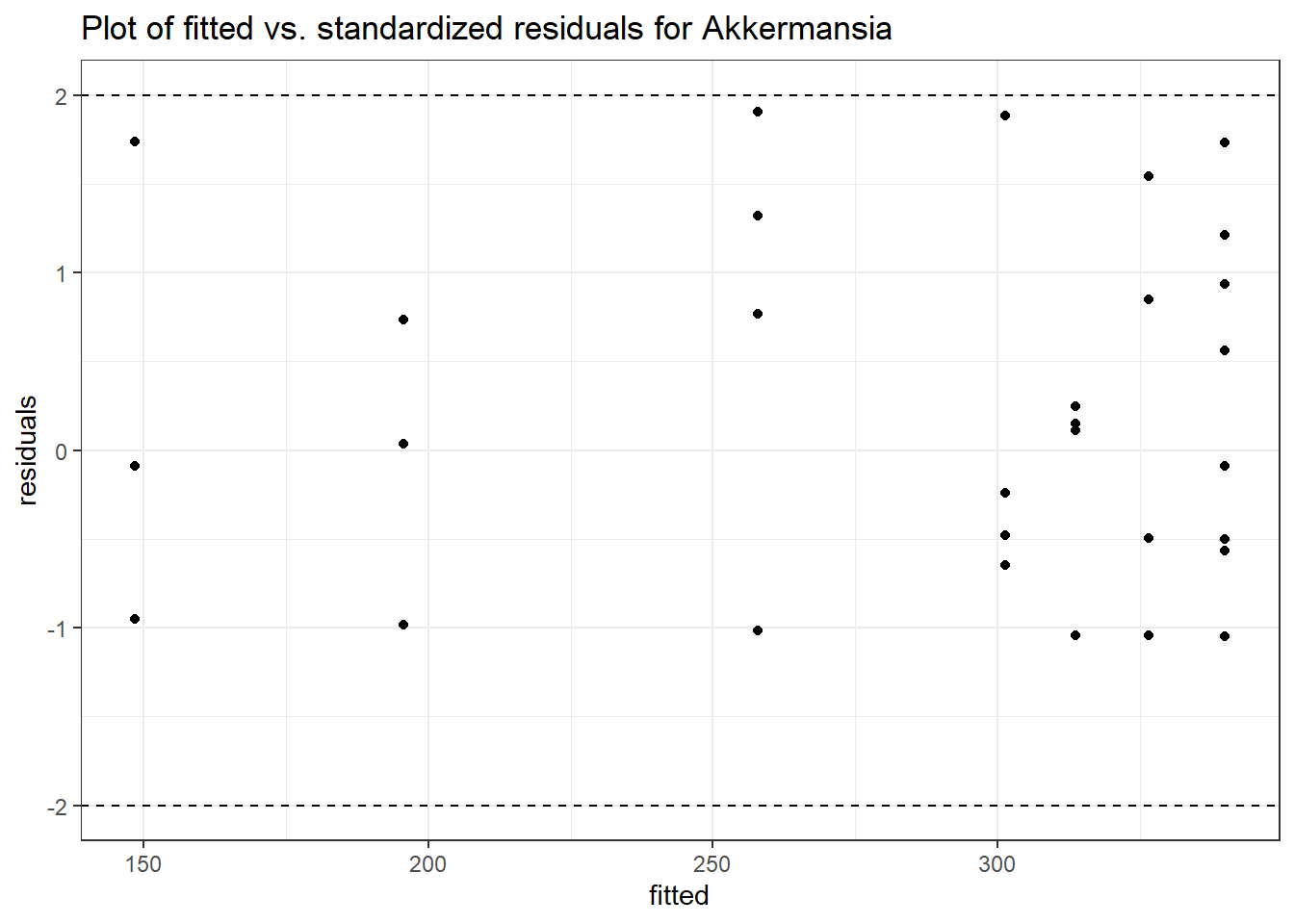
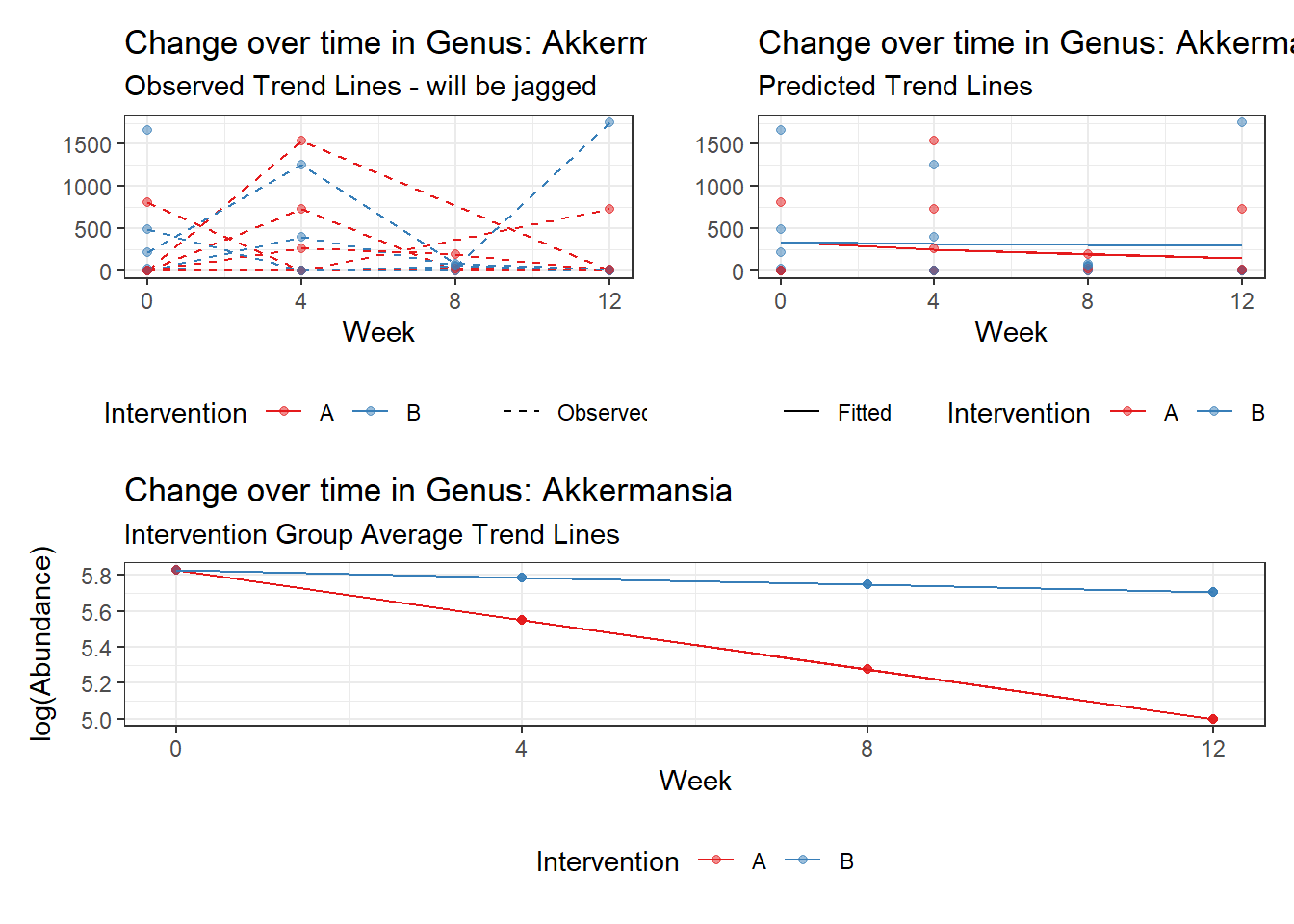
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 5.82888 0.65482 8.90148 0.00000
time Akkermansia -0.27623 0.44654 -0.61860 0.53618
time:intB Akkermansia 0.23601 0.46789 0.50442 0.61397
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.8385e-06
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinom
Intraclass Correlation (ICC): 0.2363285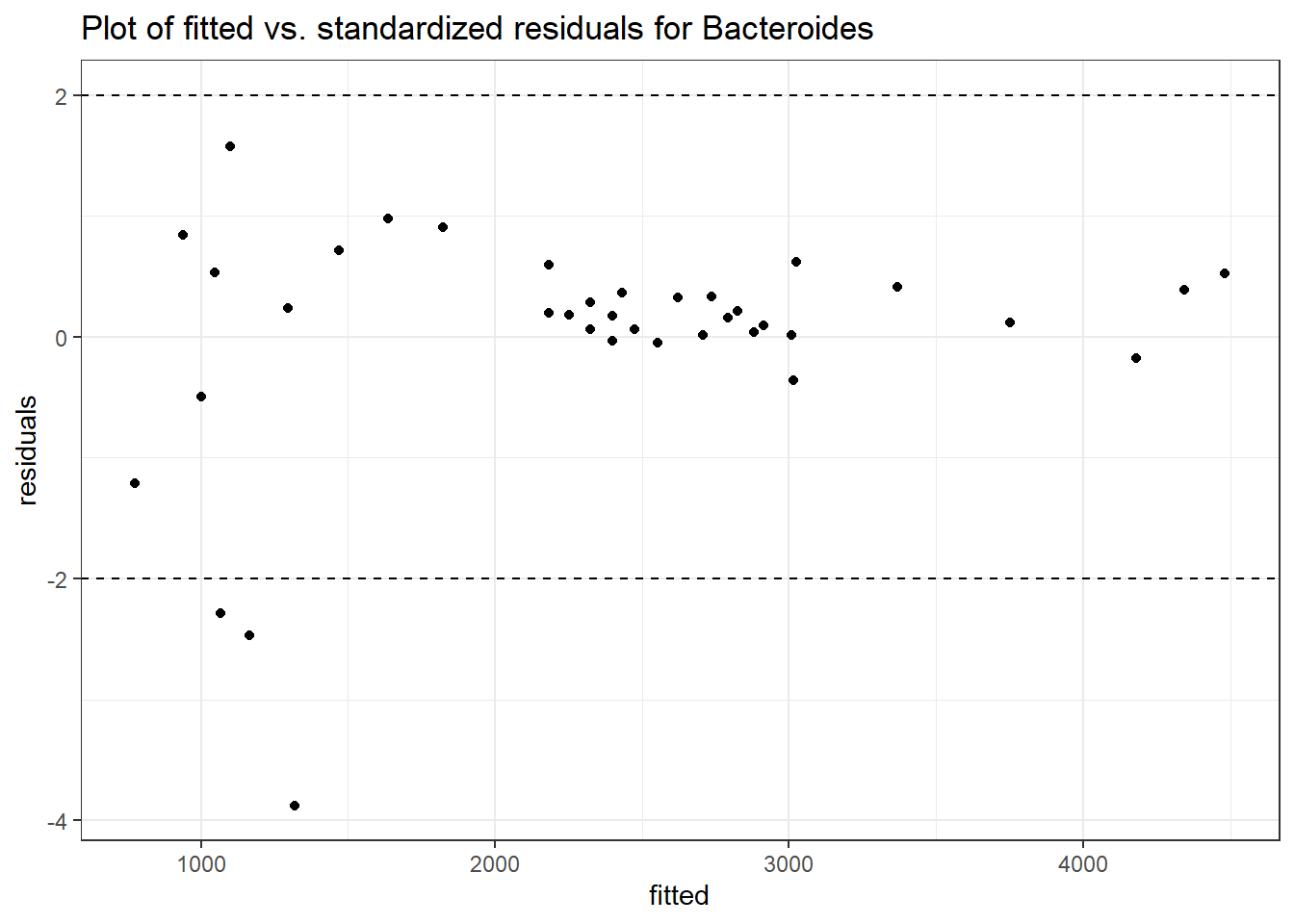
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 7.69929 0.18507 41.60297 0.00000
time Bacteroides -0.03135 0.05755 -0.54468 0.58597
time:intB Bacteroides -0.07638 0.08502 -0.89842 0.36896
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.55629
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00453326 (tol = 0.002, component 1)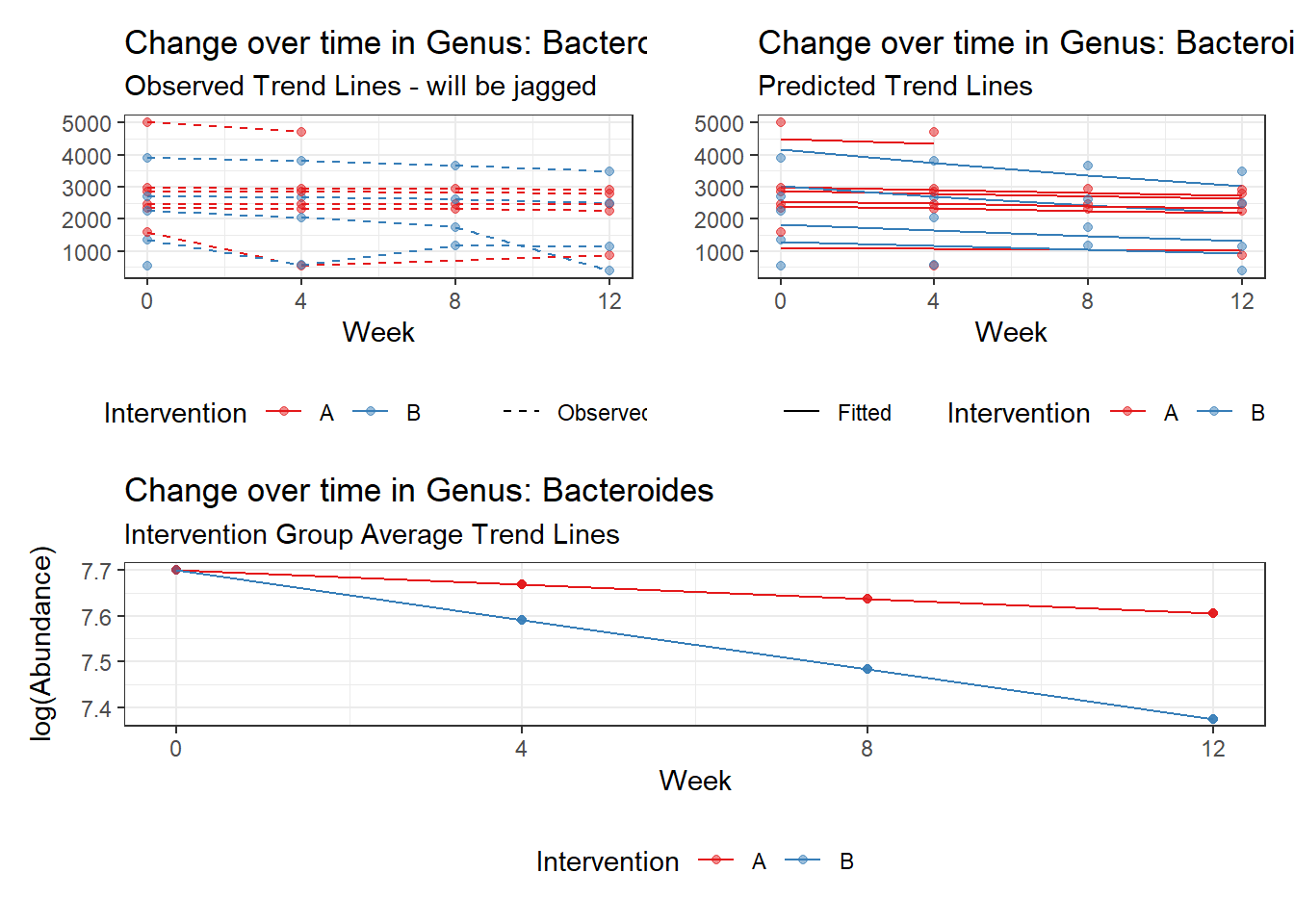
Intraclass Correlation (ICC): 0.2343535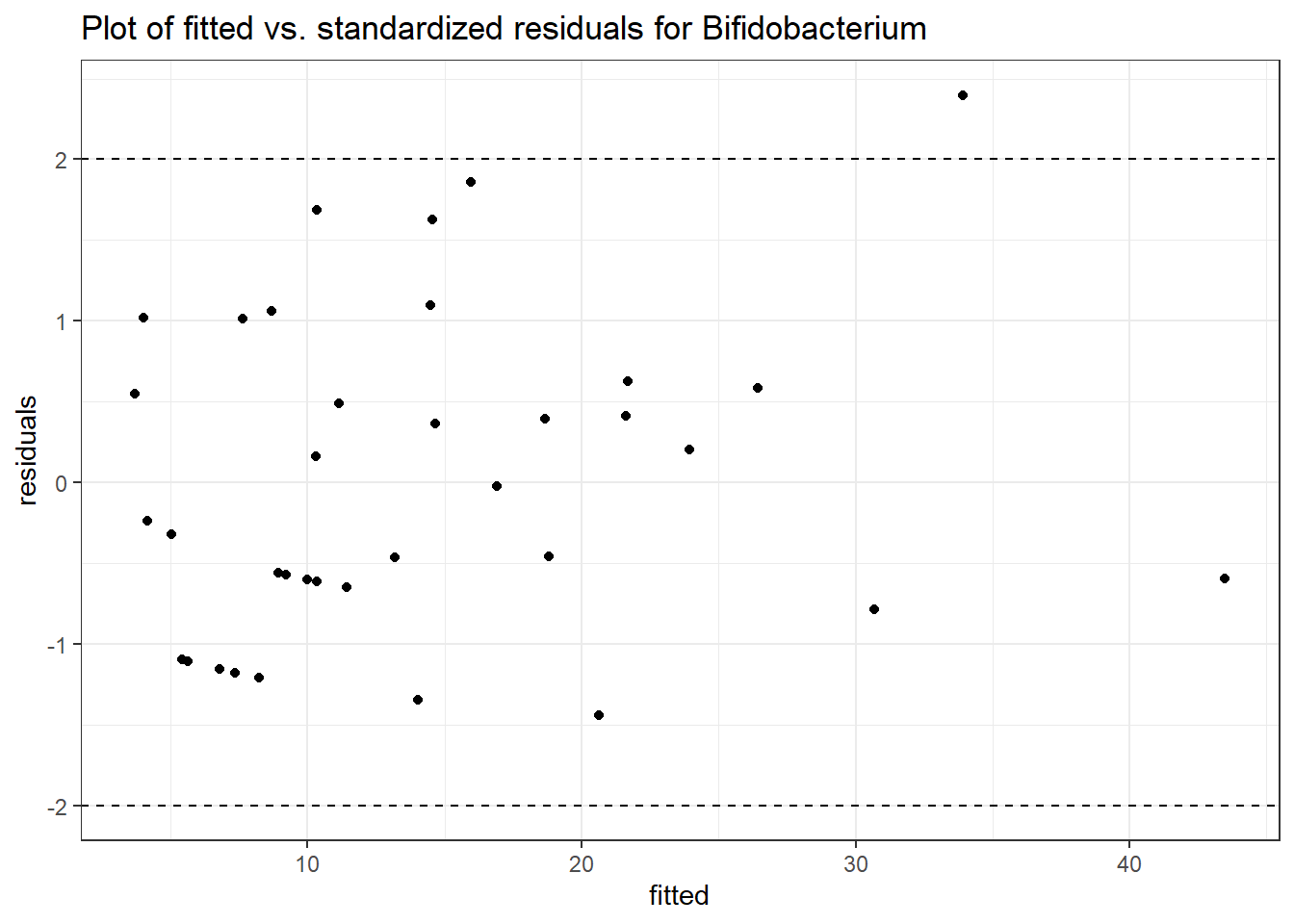
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.53313 0.54647 4.63540 0.00000
time Bifidobacterium -0.30444 0.31535 -0.96540 0.33435
time:intB Bifidobacterium 0.55311 0.36780 1.50385 0.13262
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.55325
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomboundary (singular) fit: see ?isSingular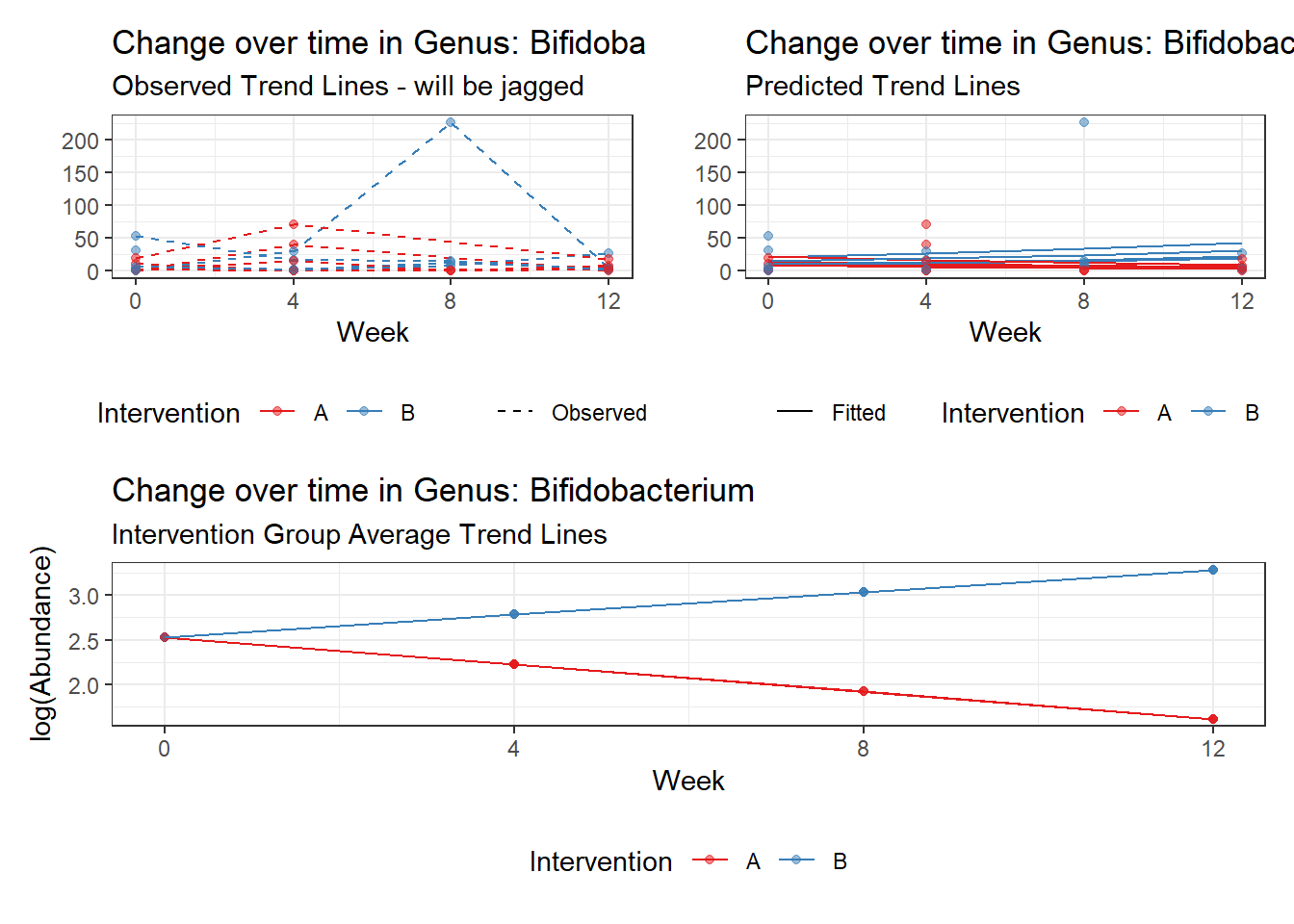
Intraclass Correlation (ICC): 6.589011e-10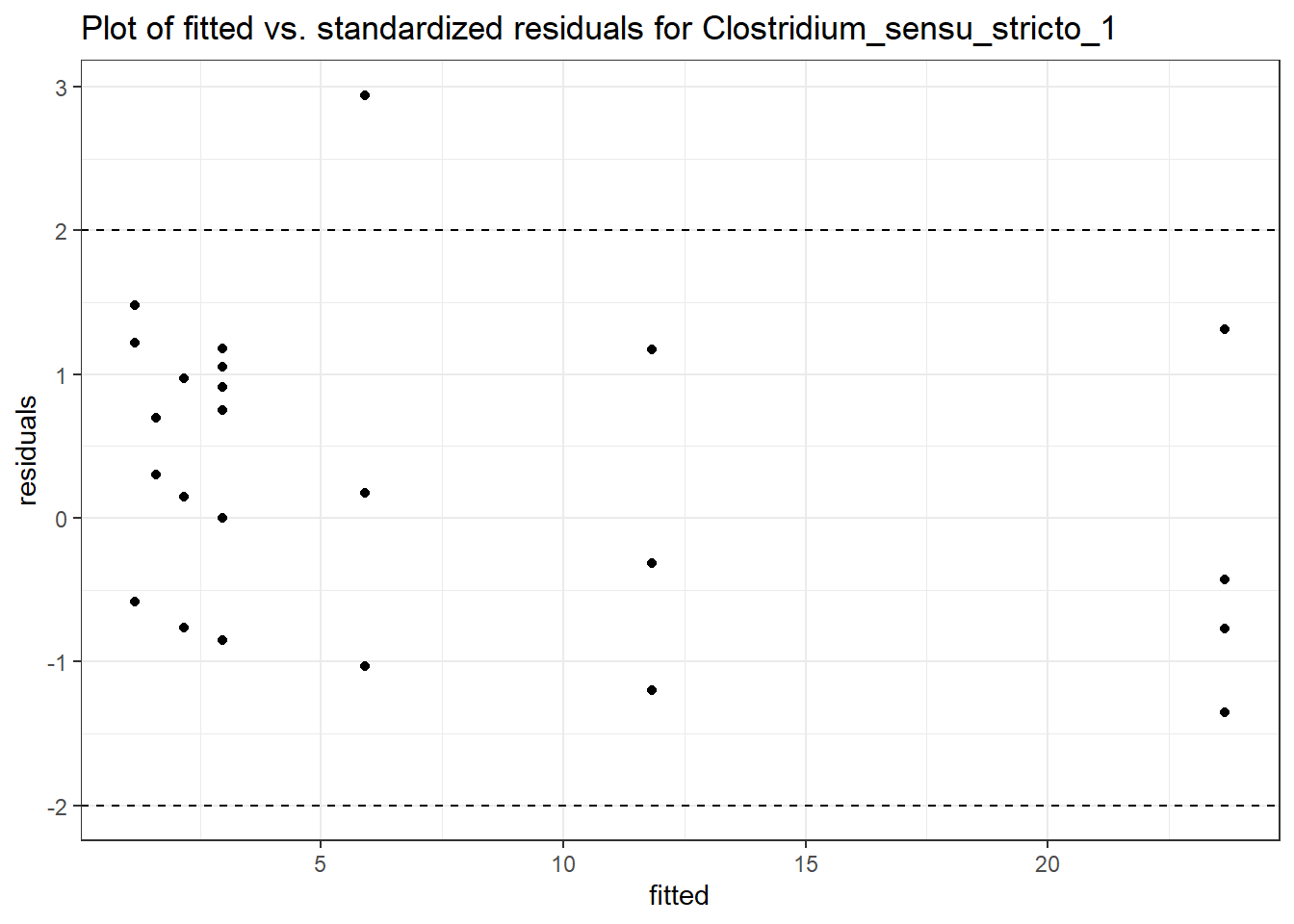

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 1.08445 0.50386 2.15229 0.03137
time Clostridium_sensu_stricto_1 -0.31134 0.31794 -0.97923 0.32747
time:intB Clostridium_sensu_stricto_1 1.00438 0.36314 2.76587 0.00568
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.5669e-05
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinom
Intraclass Correlation (ICC): 0.0566495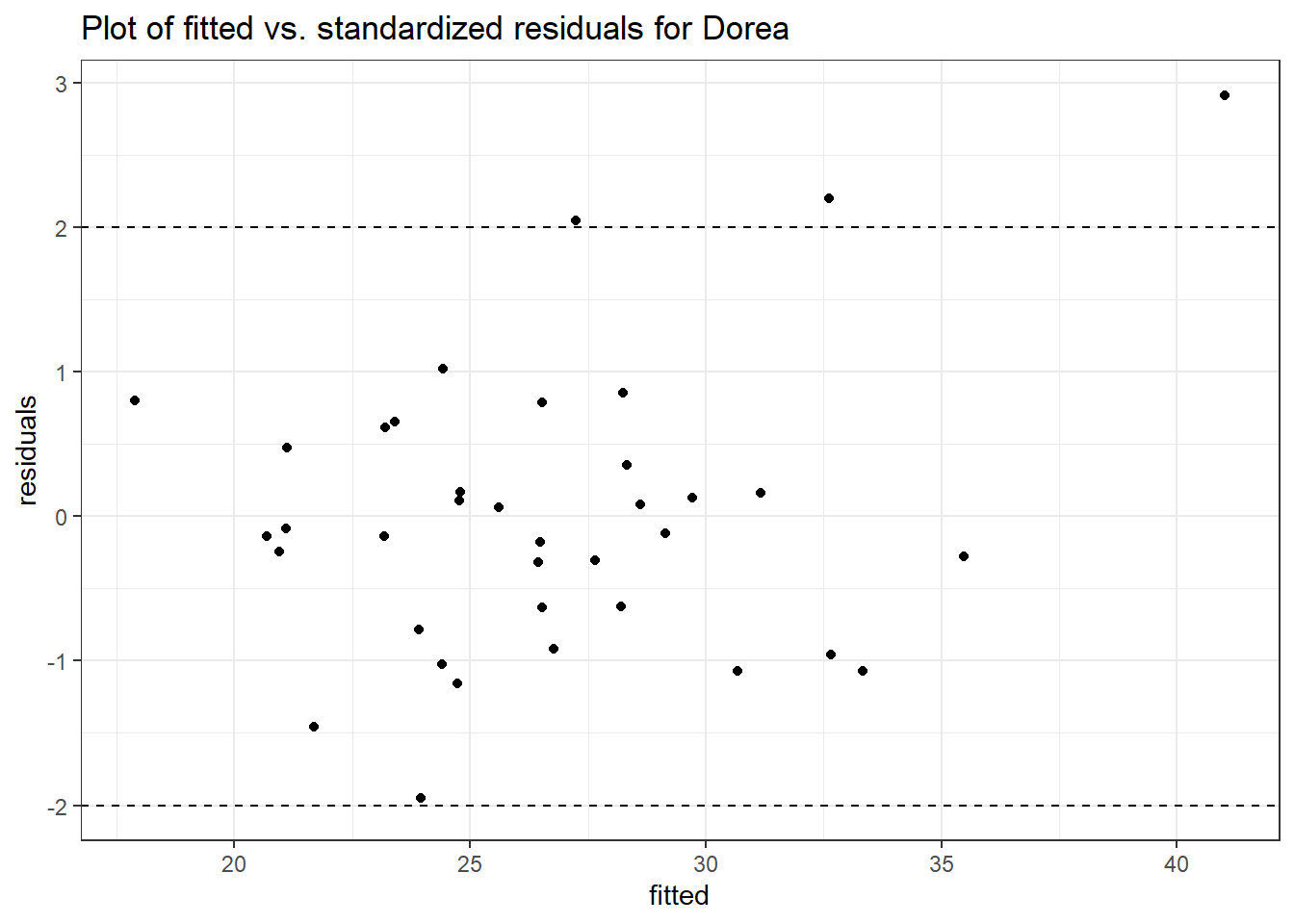
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.39921 0.20136 16.88151 0.00000
time Dorea -0.06701 0.12176 -0.55036 0.58207
time:intB Dorea -0.07815 0.14923 -0.52366 0.60051
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.24505
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 1.716444e-11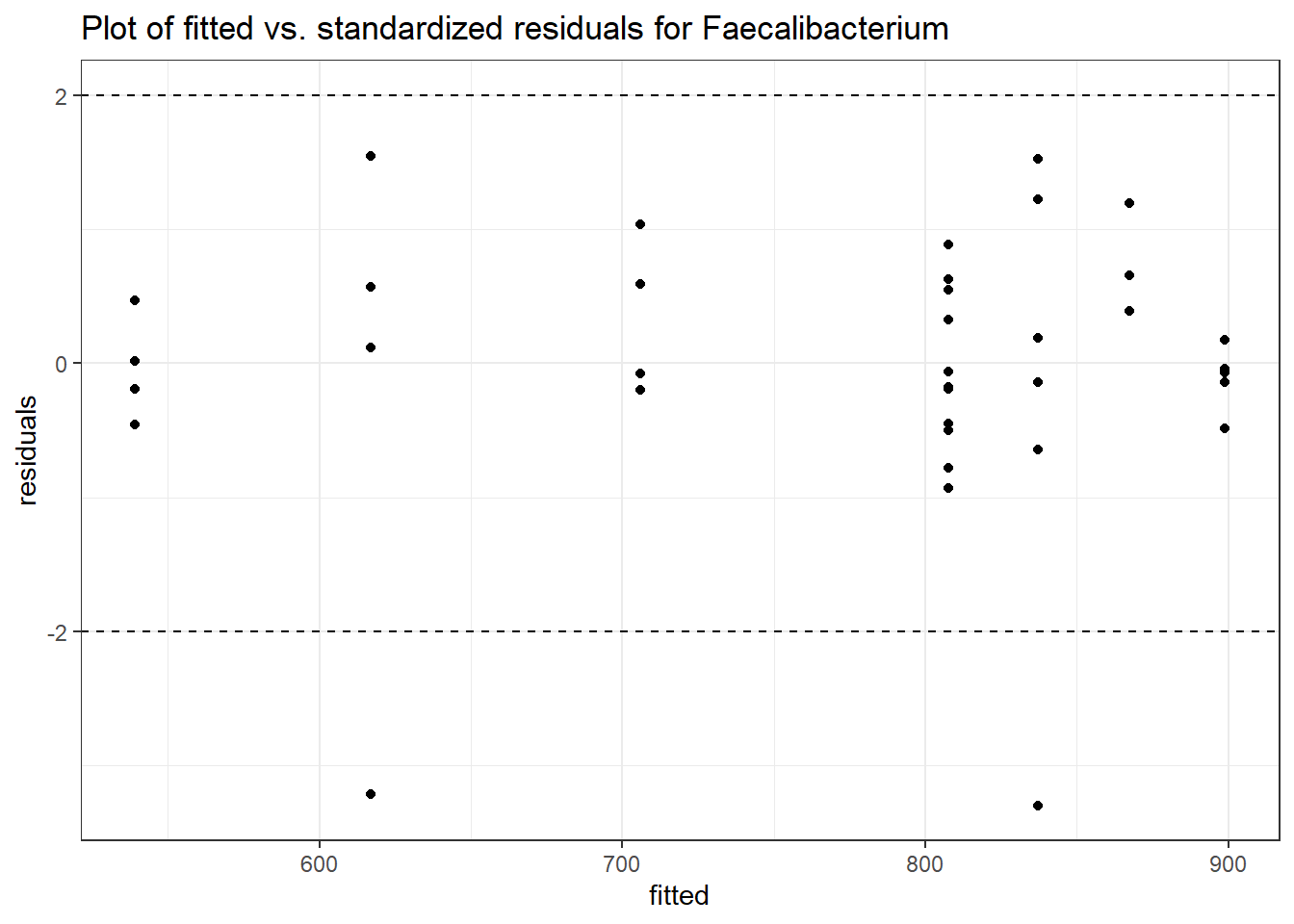
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.69428 0.26262 25.49082 0.00000
time Faecalibacterium 0.03564 0.17731 0.20099 0.84071
time:intB Faecalibacterium -0.17041 0.18284 -0.93204 0.35132
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.143e-06
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced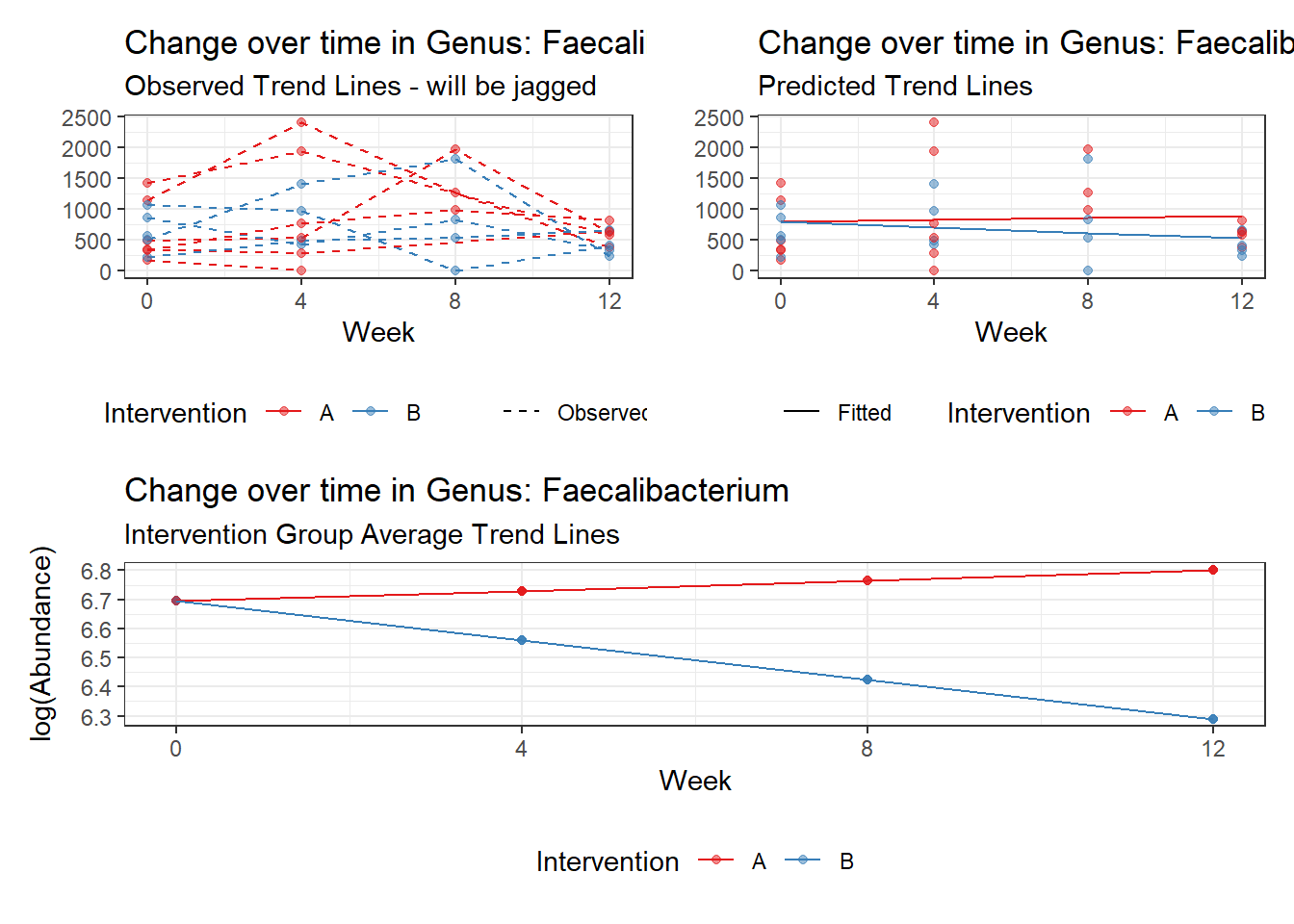
Intraclass Correlation (ICC): 0.3811174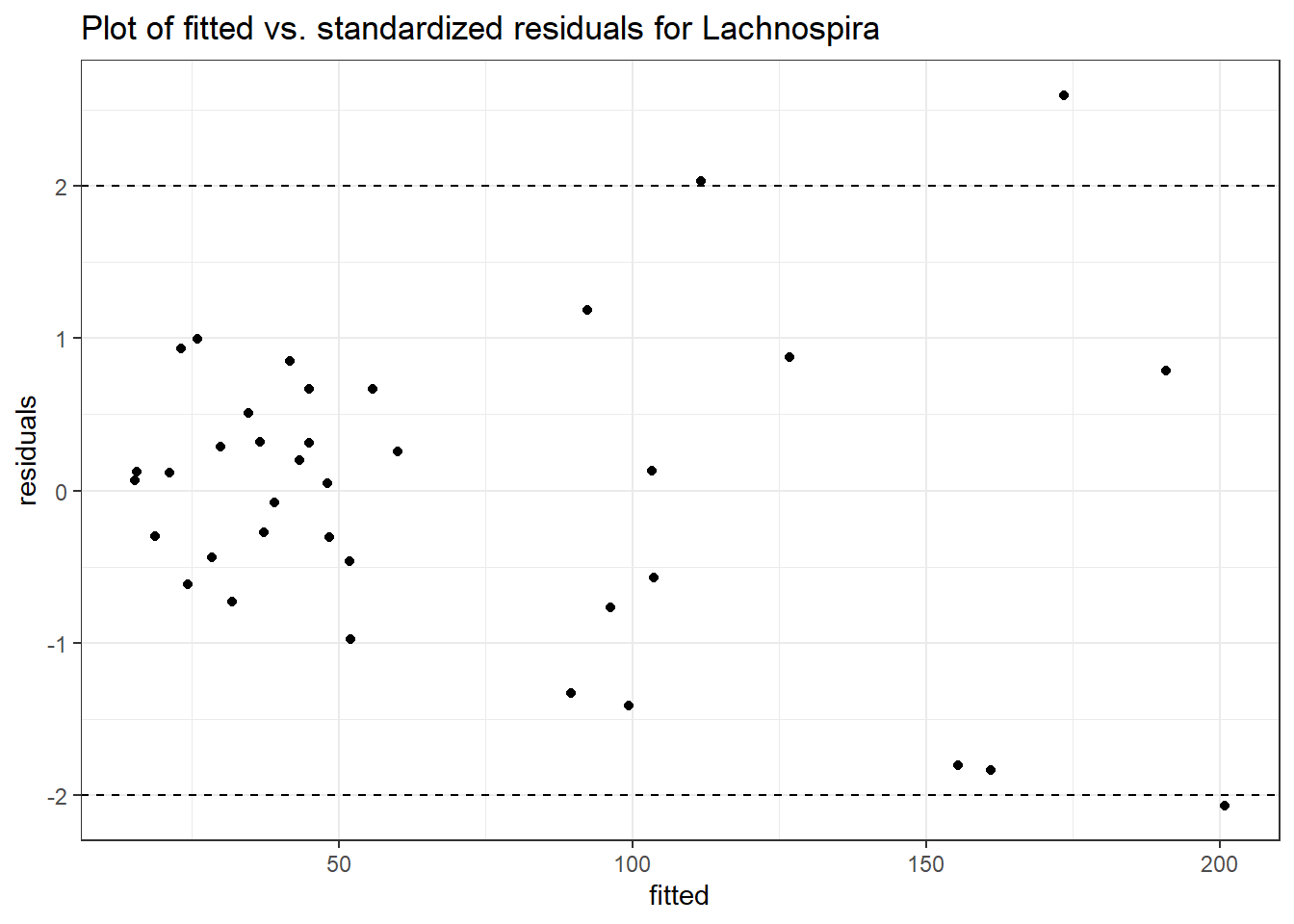
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 3.77926 0.24003 15.74482 0.00000
time Lachnospira 0.07357 0.02215 3.32102 0.00090
time:intB Lachnospira 0.13076 0.03736 3.50046 0.00046
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.78474
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomboundary (singular) fit: see ?isSingular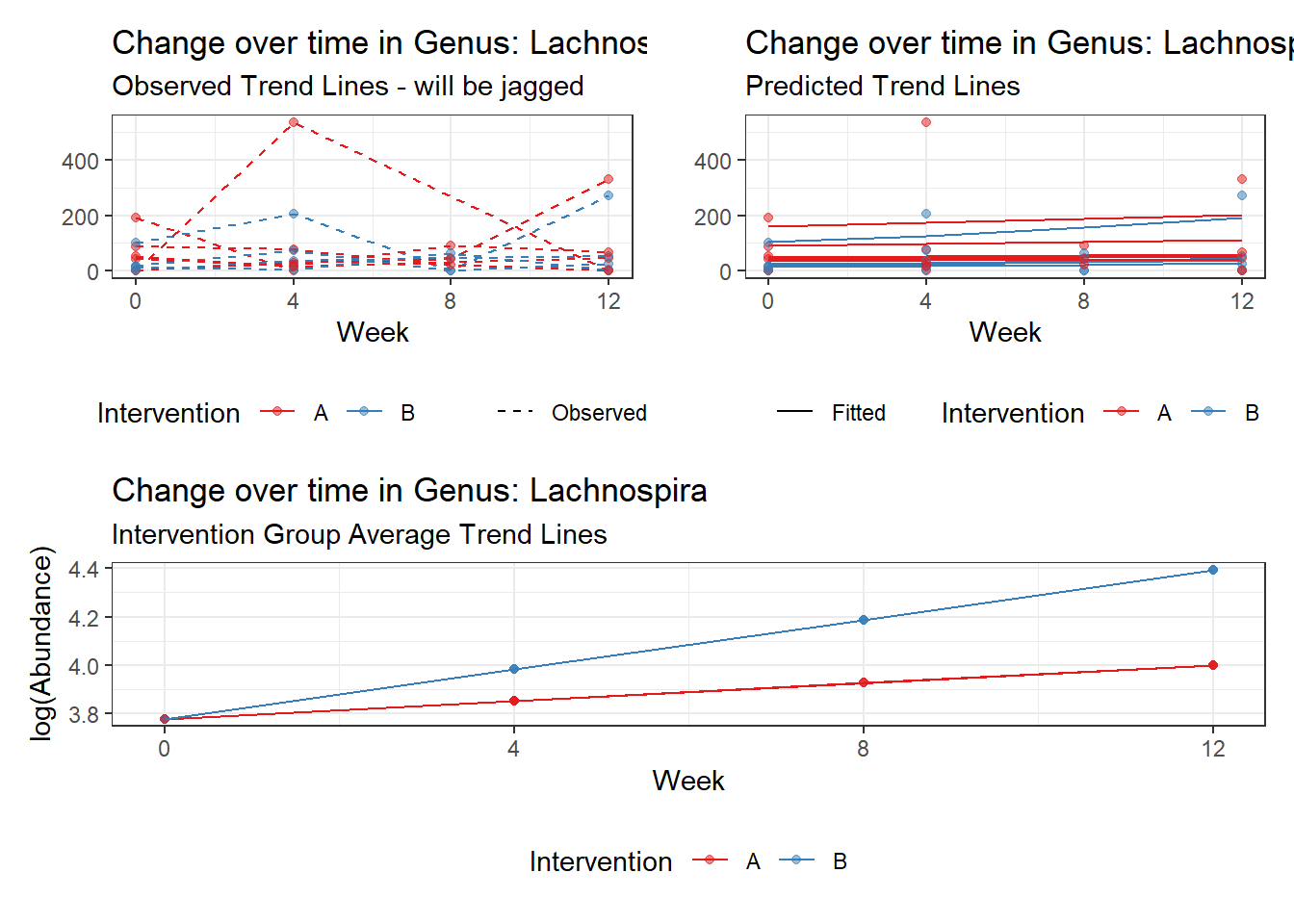
Intraclass Correlation (ICC): 5.066111e-10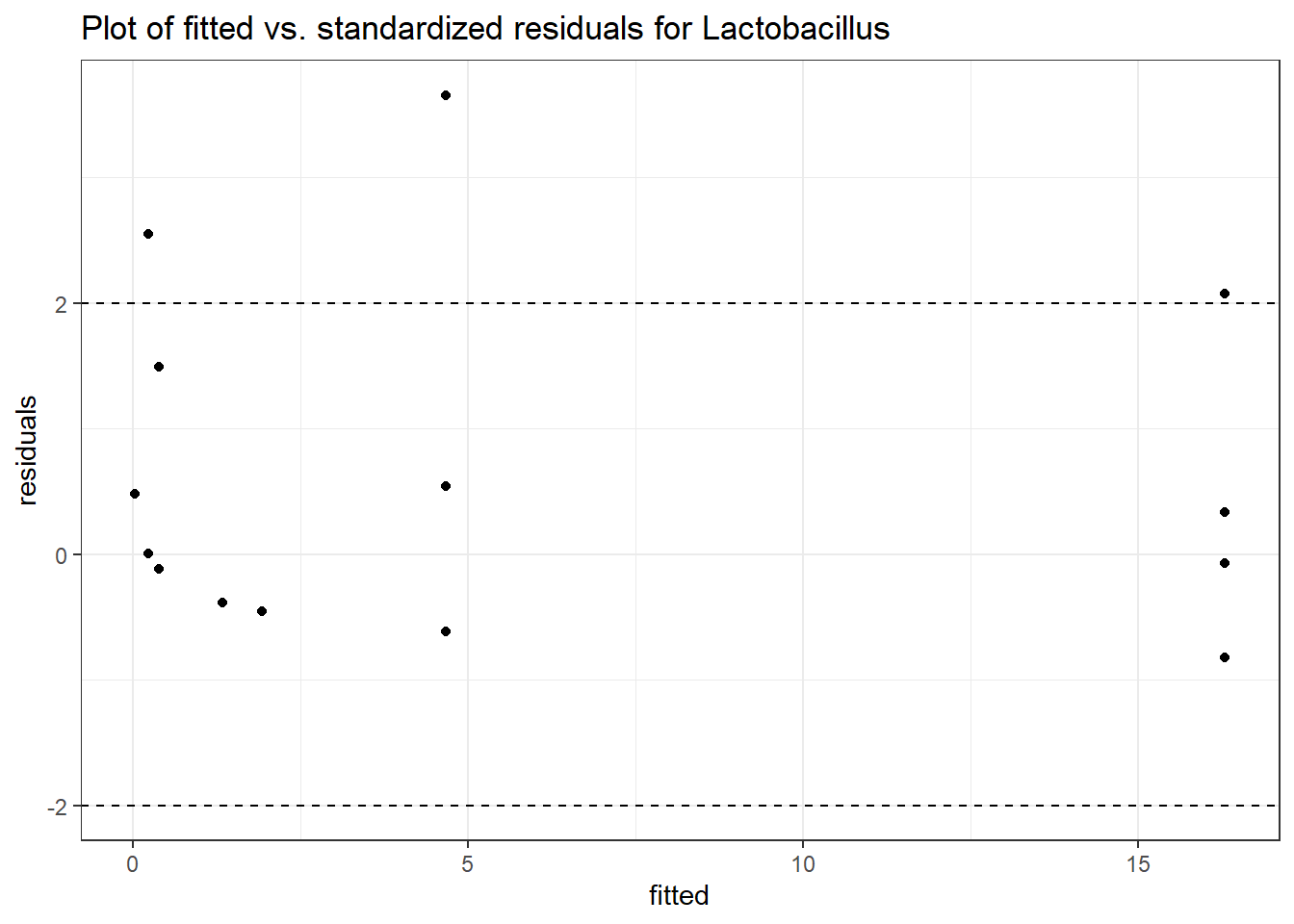
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus 2.79054 1.28437 2.17270 0.02980
time Lactobacillus -1.25202 1.01916 -1.22848 0.21927
time:intB Lactobacillus -0.88343 0.97555 -0.90557 0.36516
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.2508e-05
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00744334 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular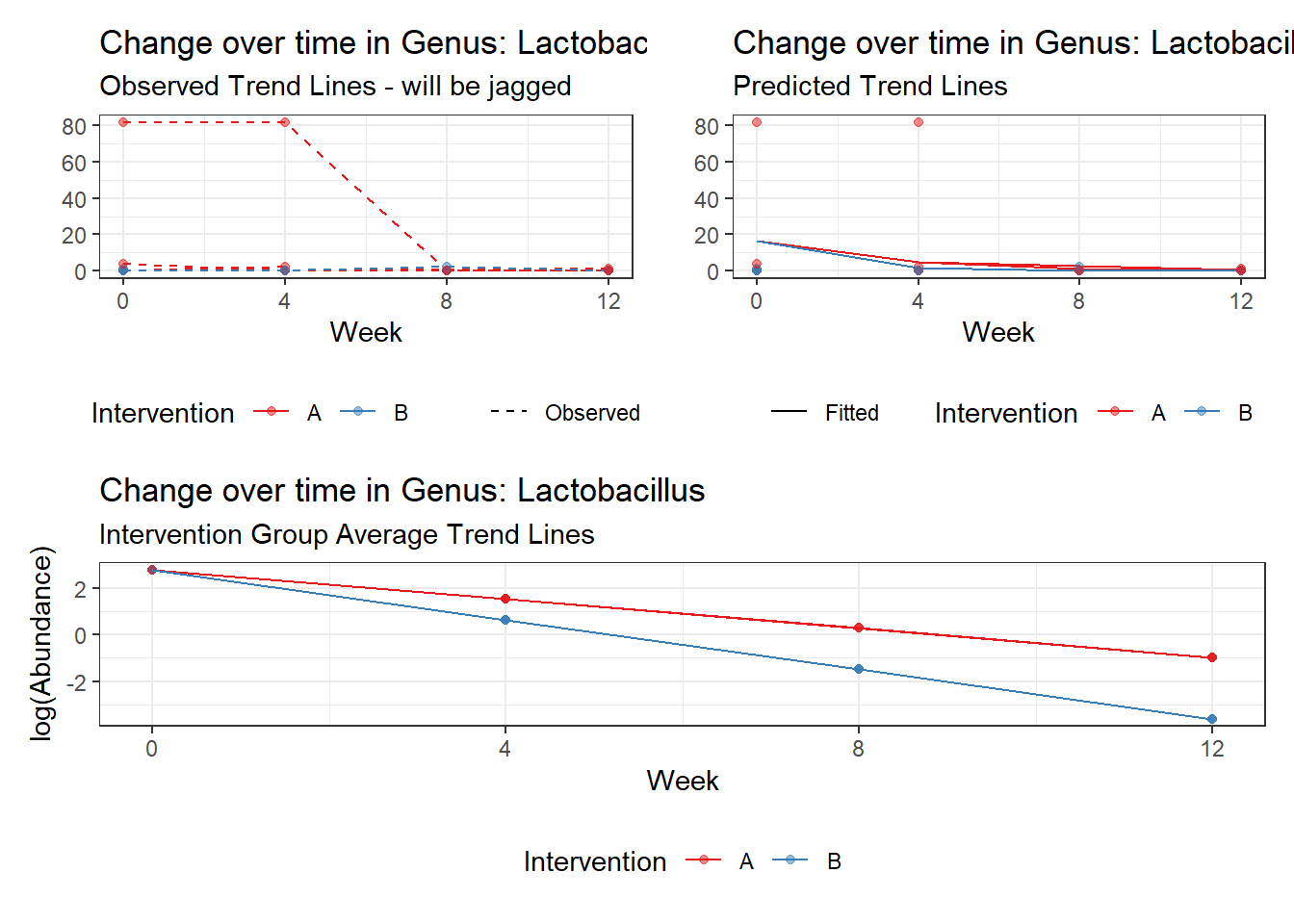
Intraclass Correlation (ICC): 9.393197e-13
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella 0.13394 0.86253 0.15529 0.87659
time Prevotella -0.06212 0.58137 -0.10685 0.91491
time:intB Prevotella 0.00335 0.61269 0.00547 0.99563
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 9.6919e-07
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 1.992685e-10
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 2.94188 0.37587 7.82685 0.00000
time Roseburia 0.14190 0.24401 0.58155 0.56087
time:intB Roseburia -0.01147 0.26970 -0.04253 0.96608
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4116e-05
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000037648d10>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 9.383701e-13

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 4.02138 0.42432 9.47727 0.00000
time Ruminococcus_1 0.29332 0.25875 1.13360 0.25696
time:intB Ruminococcus_1 -0.01805 0.30704 -0.05879 0.95312
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 9.687e-07
Significance test not available for random effectsCovariate(s): Time and Gender
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Gender)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
genus.fit7 <- glmm_microbiome(mydata=microbiome_data, model.number=7,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + time + intB:time + female + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.8231144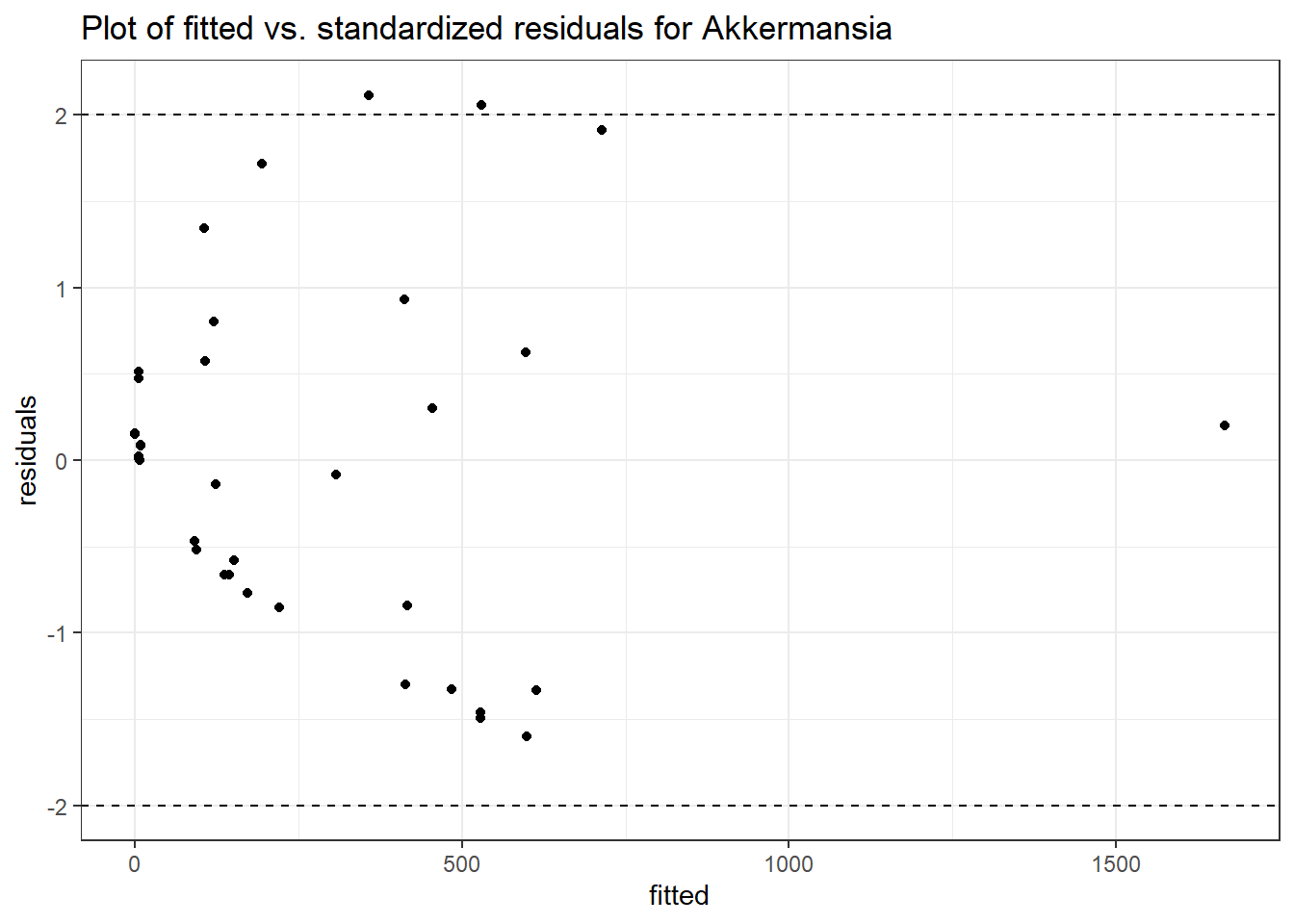
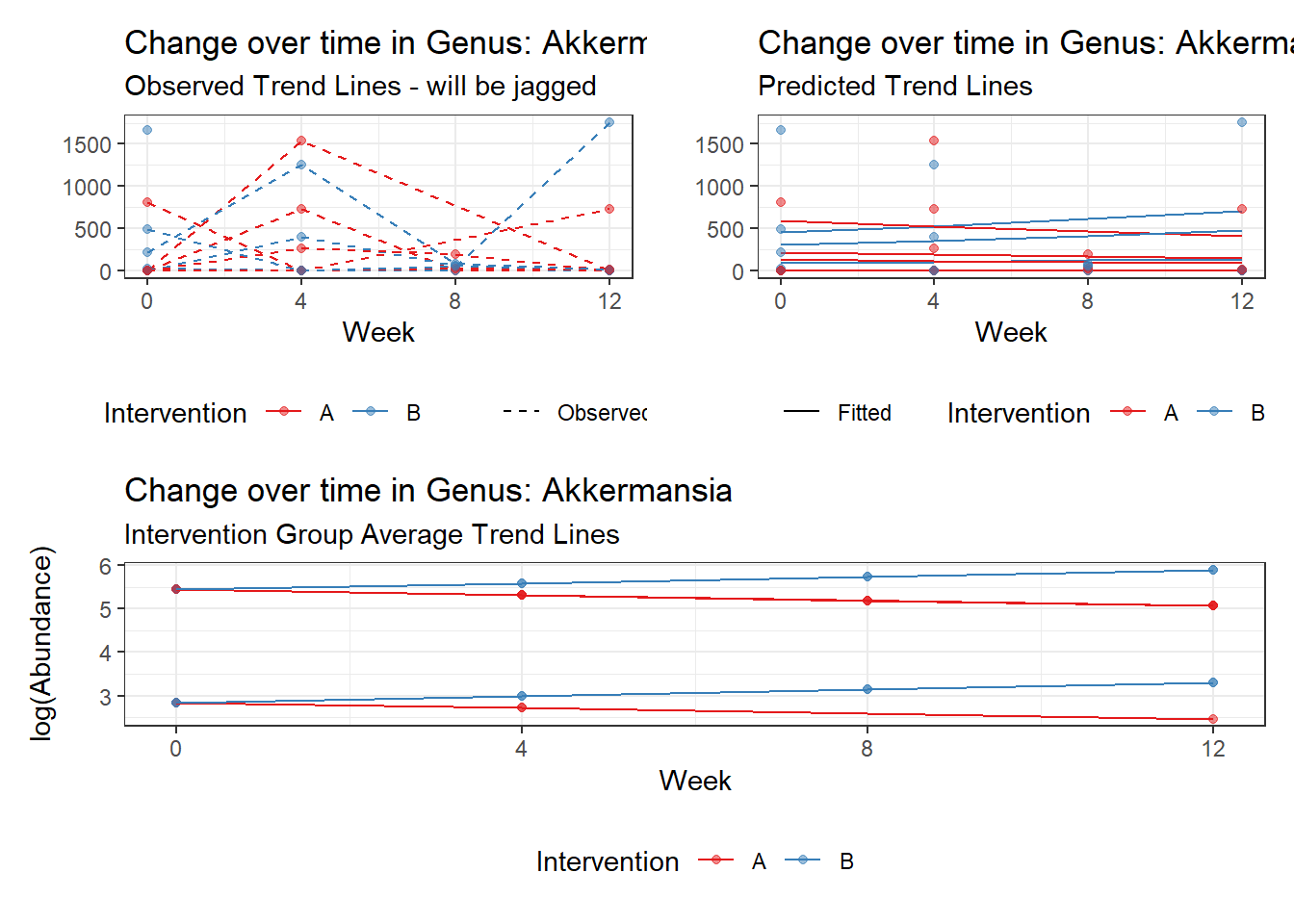
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 2.84073 1.11205 2.55450 0.01063
time Akkermansia -0.12326 0.01296 -9.50863 0.00000
female Akkermansia 2.59757 1.37874 1.88402 0.05956
time:intB Akkermansia 0.27369 0.01882 14.53922 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1572
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.180896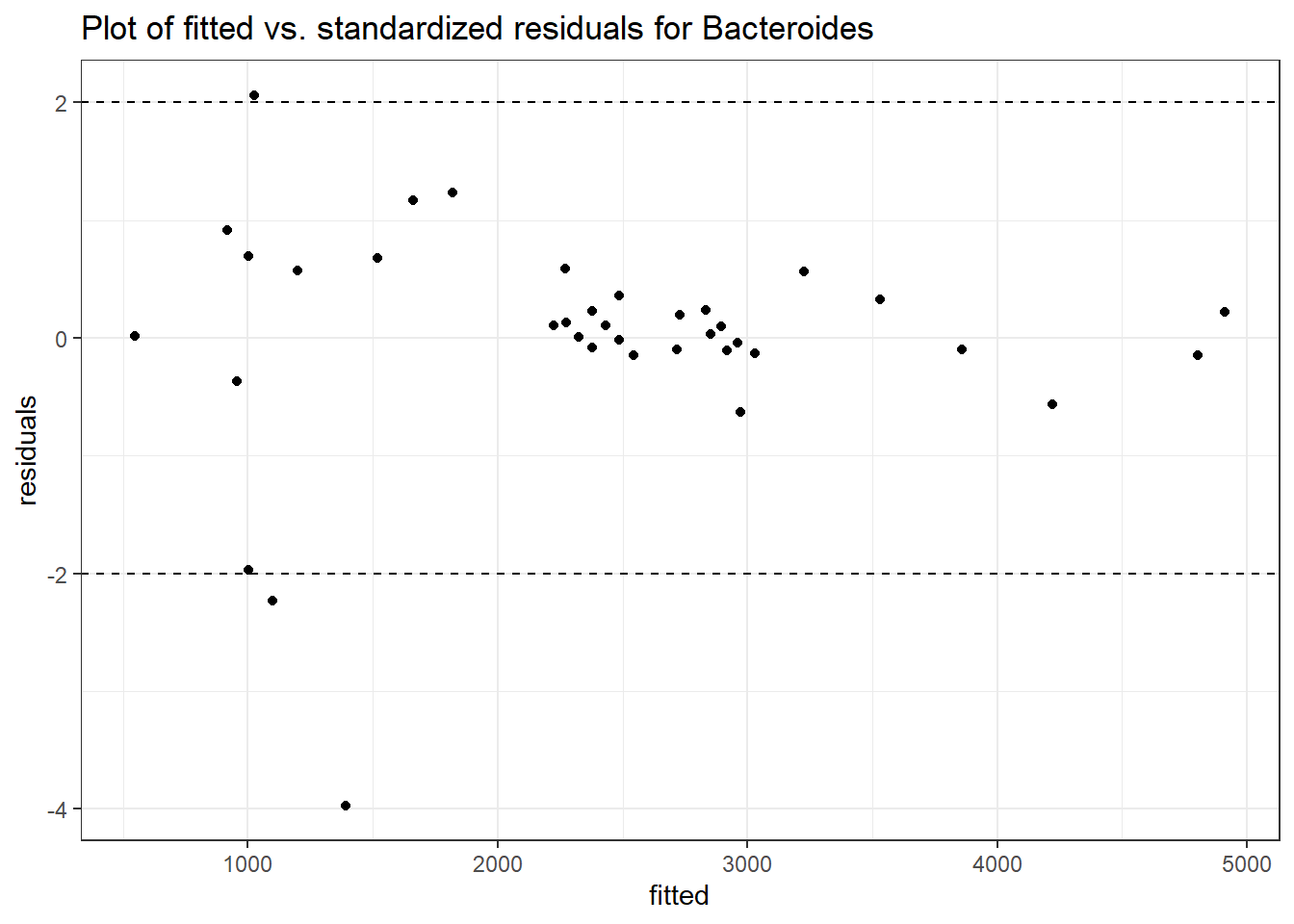
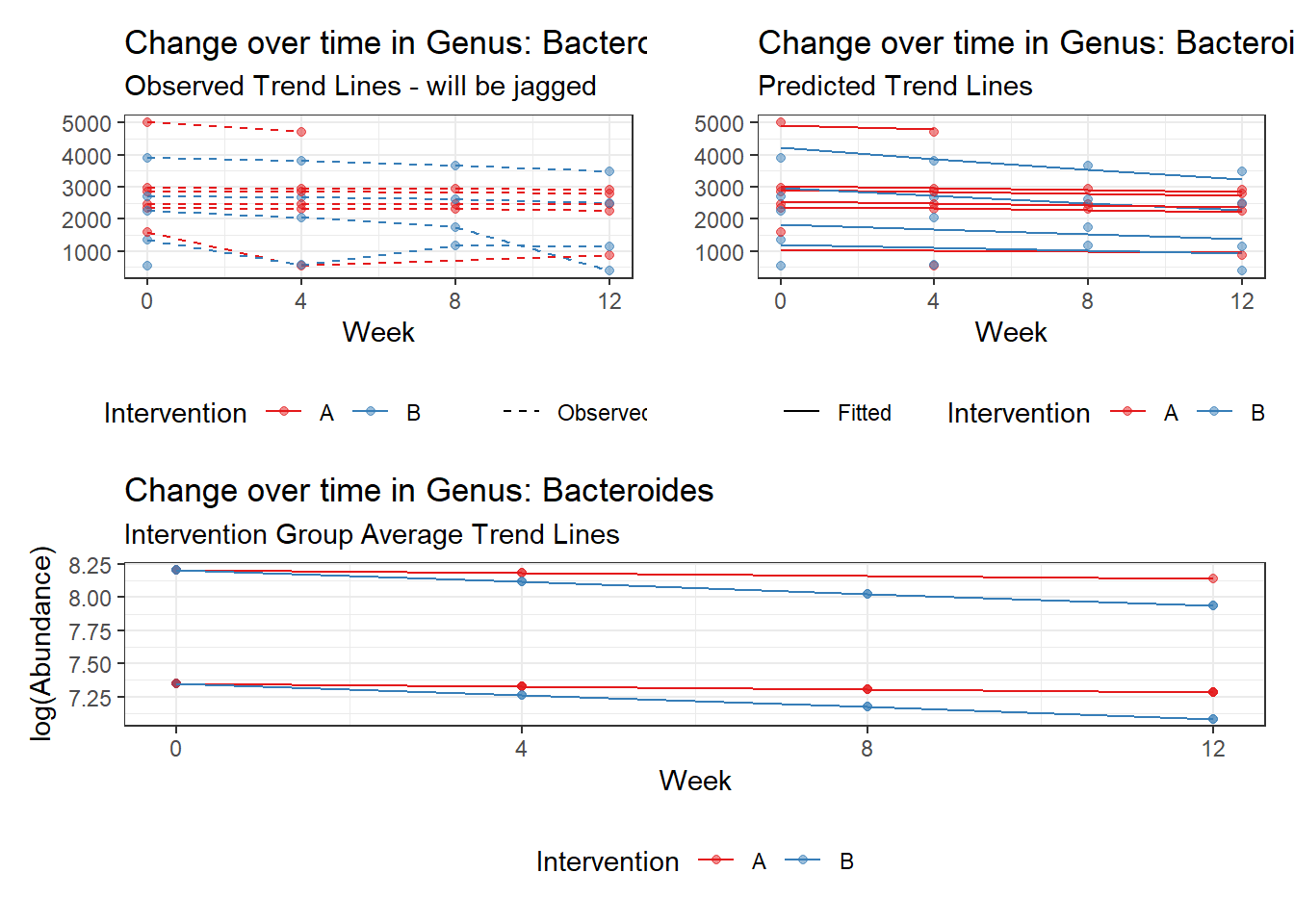
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 8.20579 0.23505 34.91083 0.00000
time Bacteroides -0.02244 0.00413 -5.42888 0.00000
female Bacteroides -0.85386 0.29470 -2.89744 0.00376
time:intB Bacteroides -0.06719 0.00628 -10.69146 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.46994
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.5362481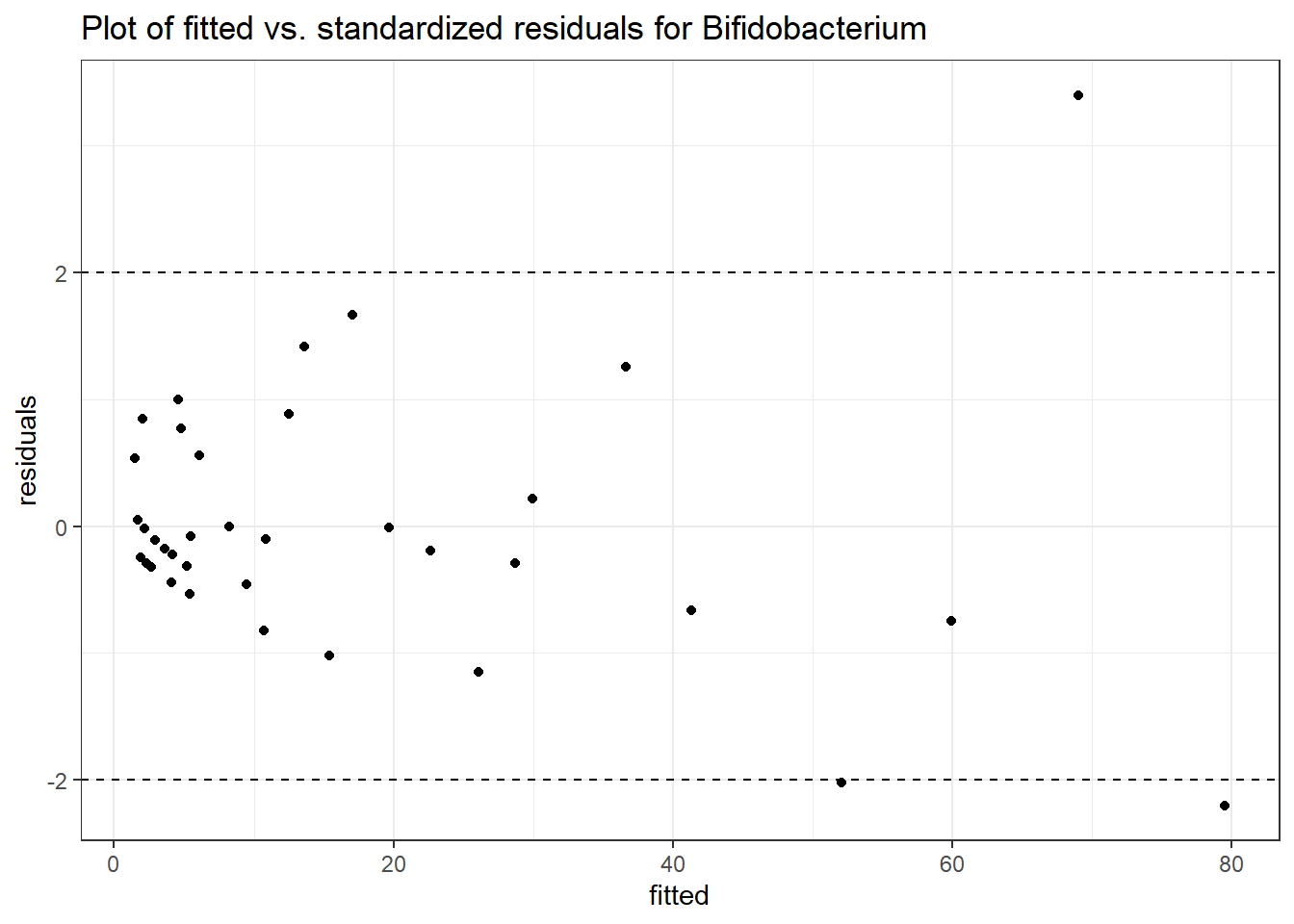

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.42492 0.54931 4.41446 0.00001
time Bifidobacterium -0.12098 0.06173 -1.95978 0.05002
female Bifidobacterium -0.22485 0.68762 -0.32700 0.74367
time:intB Bifidobacterium 0.26222 0.07581 3.45903 0.00054
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0753
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + intB:time + female +
(1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.5970951
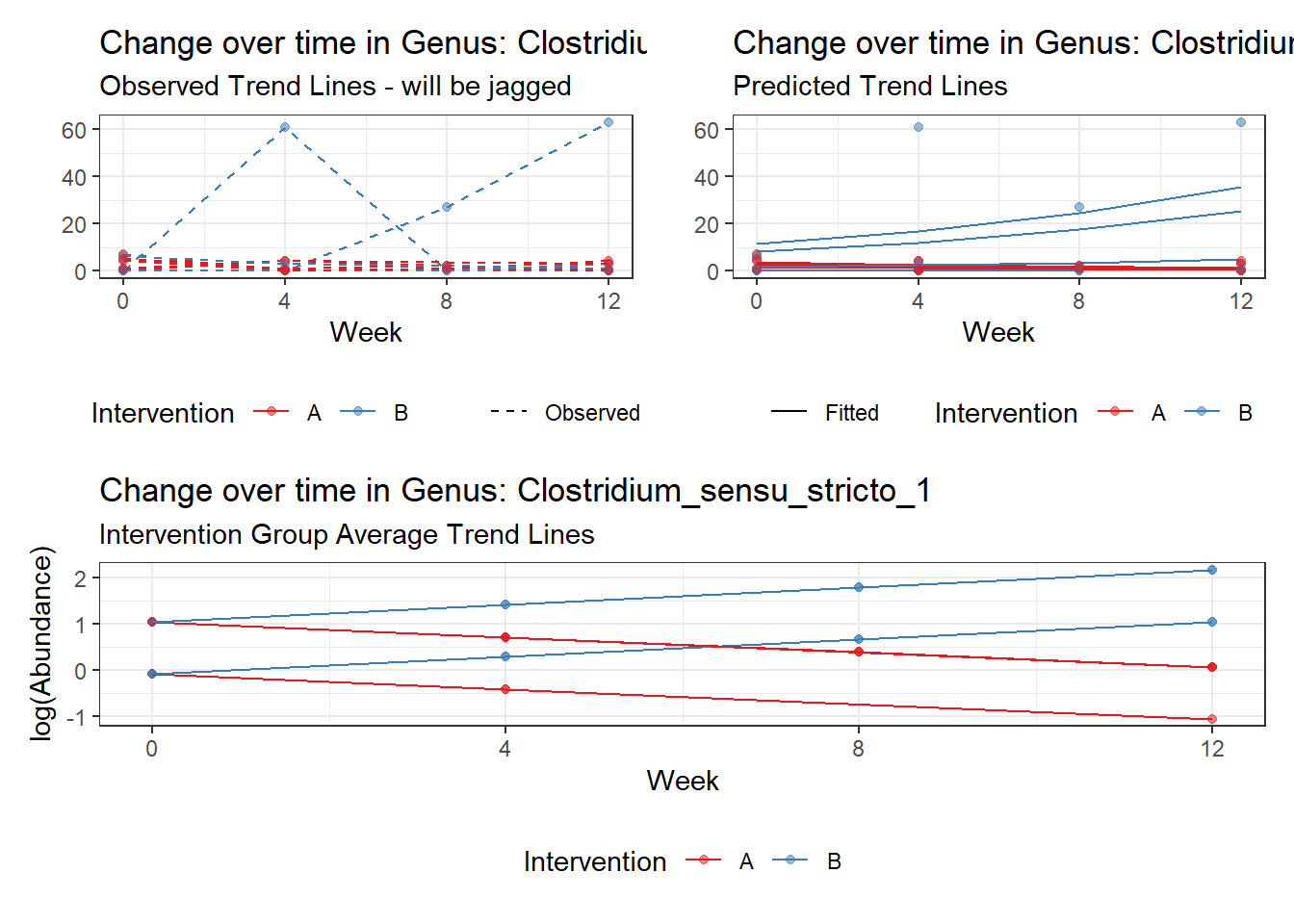
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 -0.08638 0.74301 -0.11625 0.90745
time Clostridium_sensu_stricto_1 -0.32471 0.14723 -2.20547 0.02742
female Clostridium_sensu_stricto_1 1.12902 0.87720 1.28707 0.19807
time:intB Clostridium_sensu_stricto_1 0.70239 0.16254 4.32130 0.00002
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.2174
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.2721939
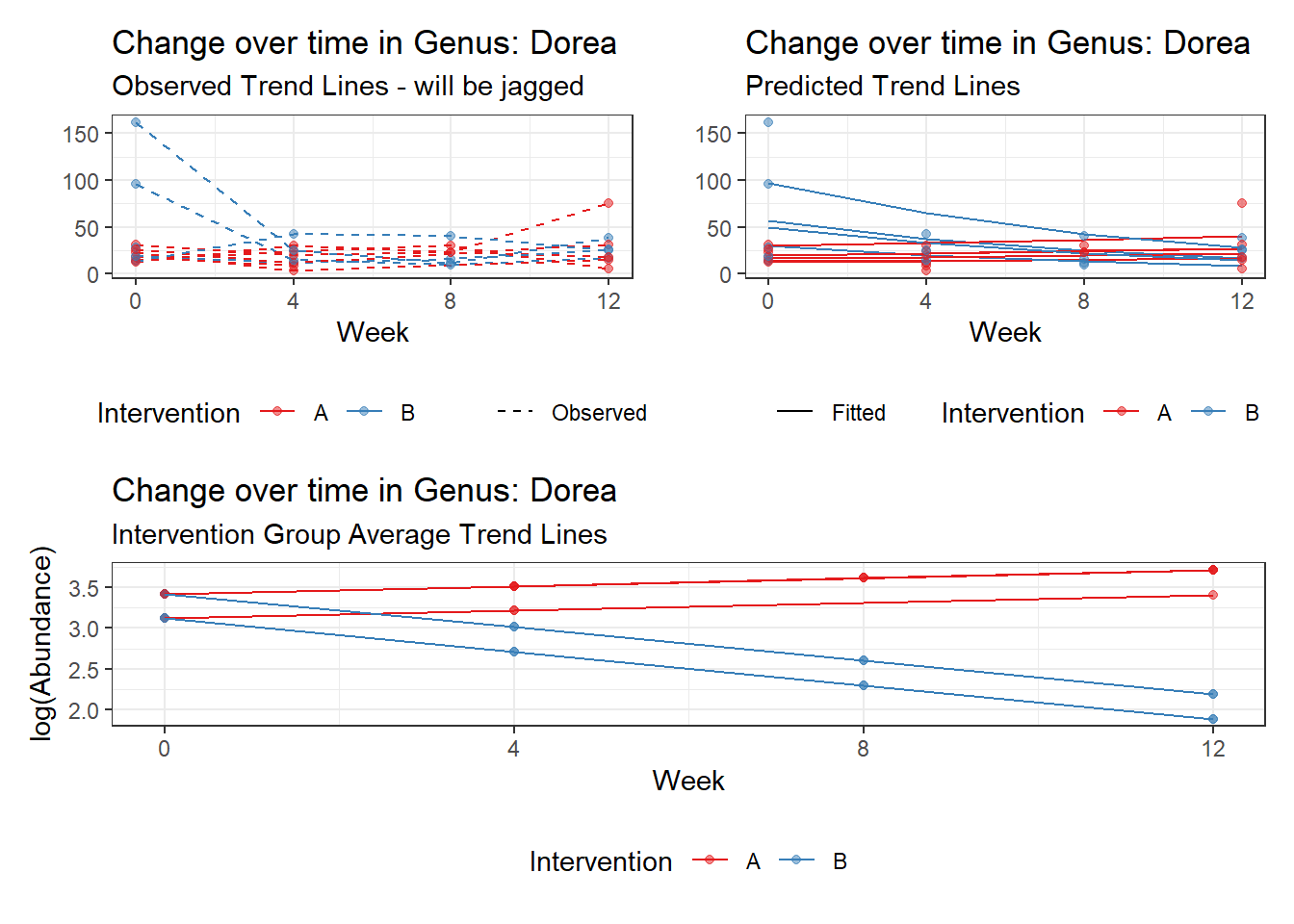
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.11803 0.31533 9.88830 0.00000
time Dorea 0.09730 0.04190 2.32205 0.02023
female Dorea 0.30394 0.39267 0.77405 0.43890
time:intB Dorea -0.50783 0.05868 -8.65346 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.61155
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.3249697
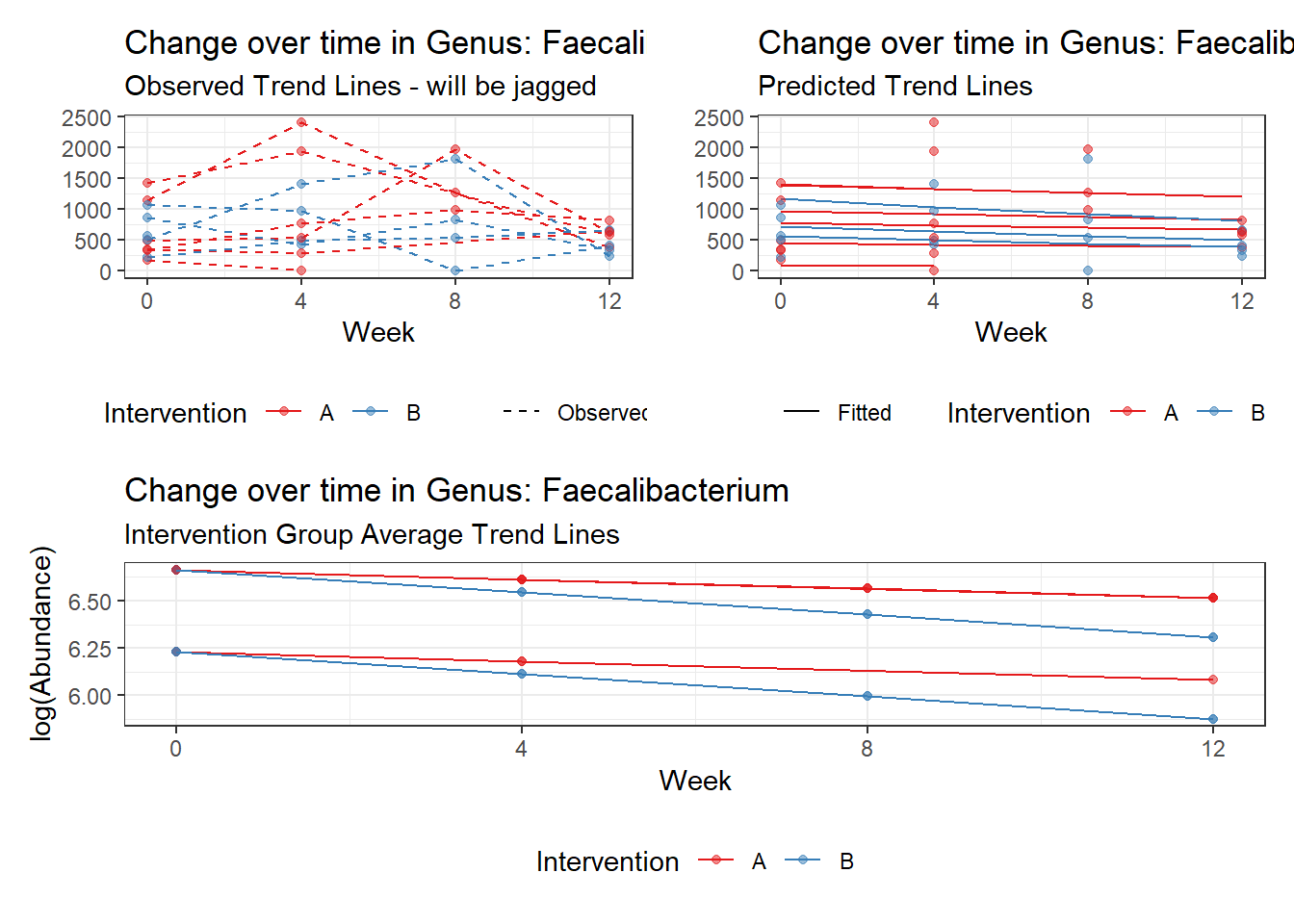
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.23061 0.34757 17.92611 0.00000
time Faecalibacterium -0.04940 0.00665 -7.42726 0.00000
female Faecalibacterium 0.43166 0.43546 0.99127 0.32155
time:intB Faecalibacterium -0.06852 0.01094 -6.26607 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.69384
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.2948803
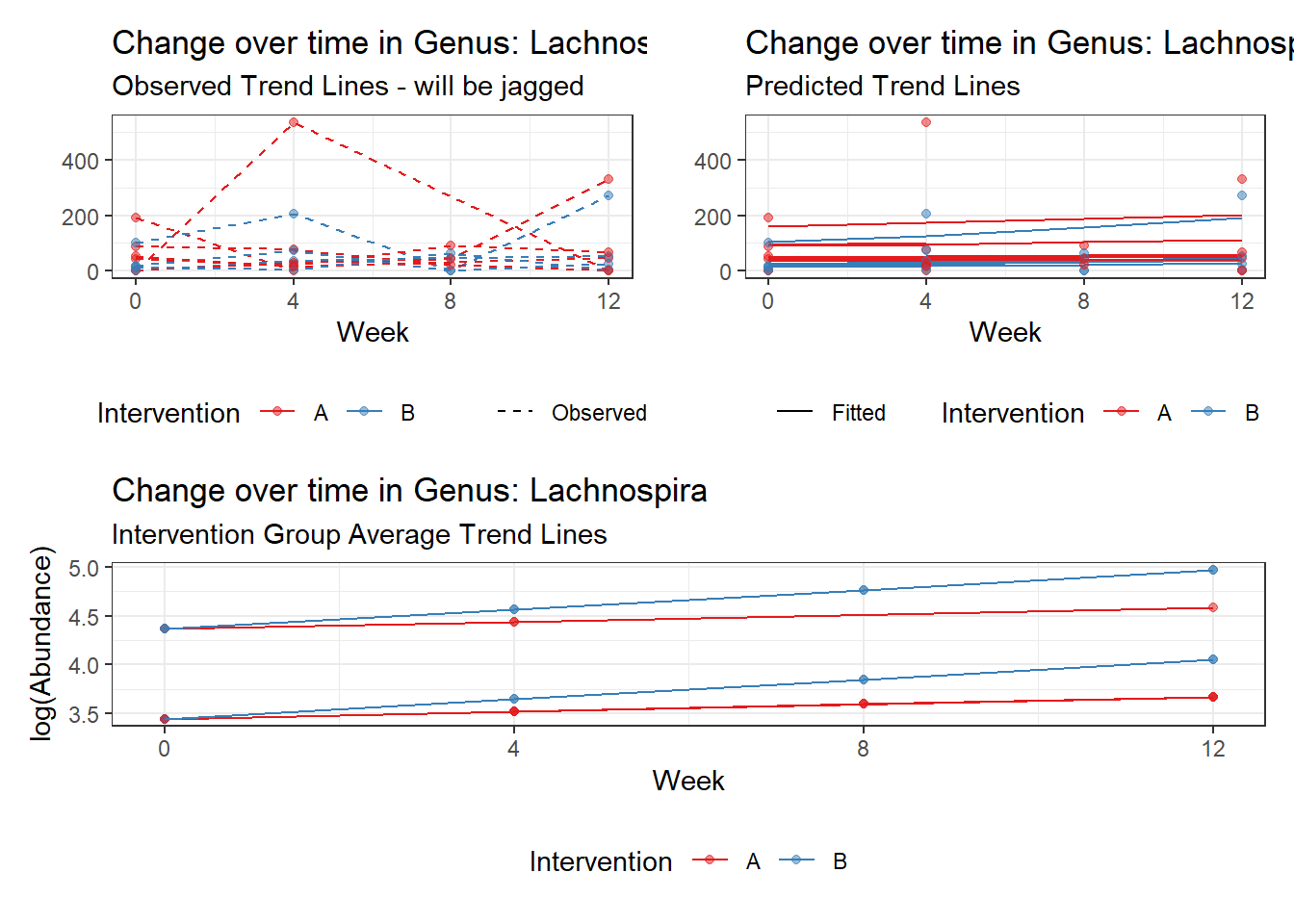
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.36472 0.32612 13.38378 0.00000
time Lachnospira 0.07427 0.02190 3.39198 0.00069
female Lachnospira -0.91773 0.40930 -2.24220 0.02495
time:intB Lachnospira 0.12730 0.03712 3.42901 0.00061
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.64668
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.8721755
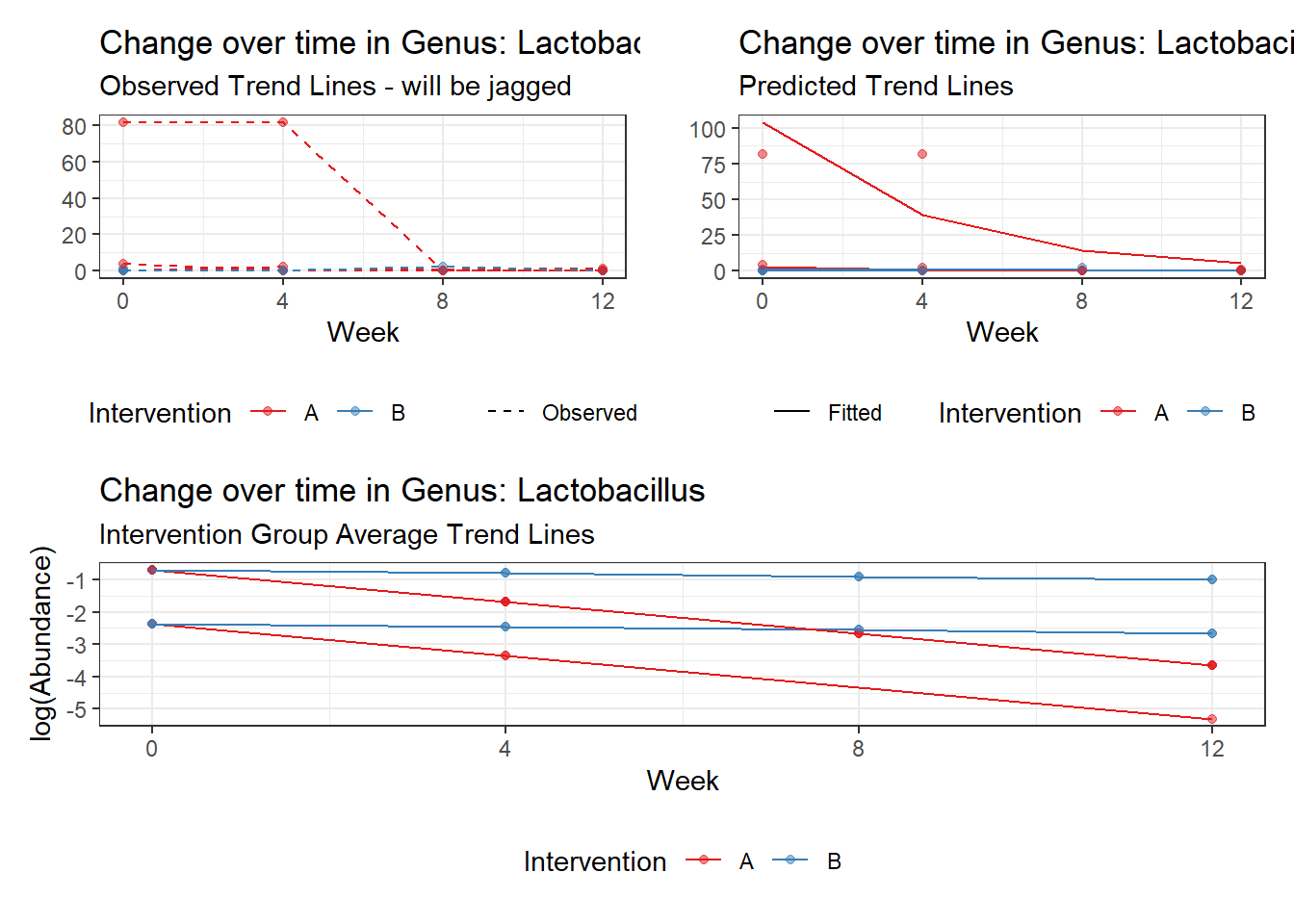
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus -2.35252 1.73482 -1.35607 0.17508
time Lactobacillus -0.98331 0.09737 -10.09914 0.00000
female Lactobacillus 1.65263 1.95363 0.84592 0.39759
time:intB Lactobacillus 0.88275 0.52325 1.68707 0.09159
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6121
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.7582225

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.54157 1.15544 -1.33418 0.18214
time Prevotella 0.11667 0.18722 0.62320 0.53315
female Prevotella 0.83861 1.30869 0.64080 0.52165
time:intB Prevotella -0.03576 0.28377 -0.12601 0.89972
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7709
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.6914695
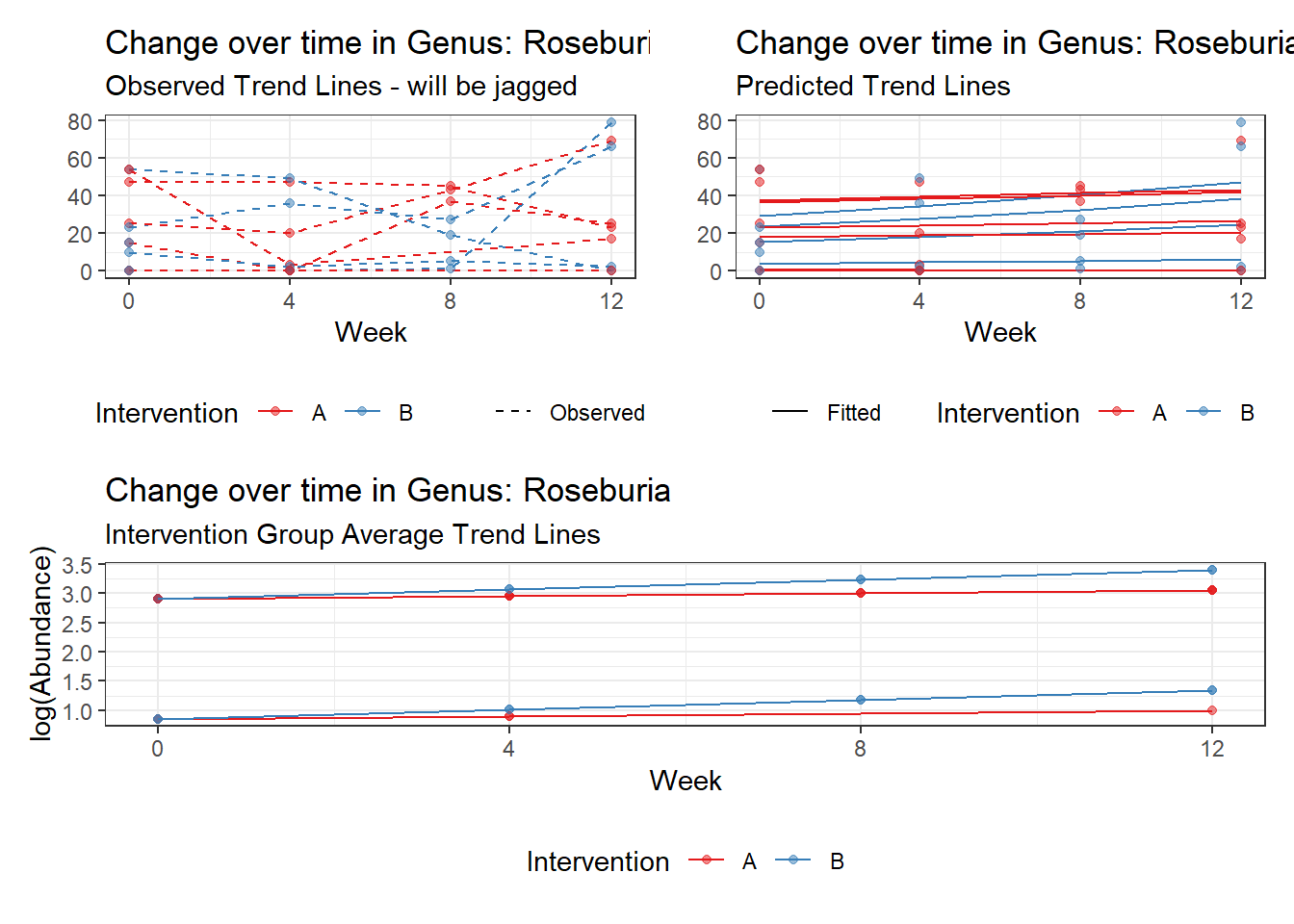
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 0.85787 0.85302 1.00568 0.31457
time Roseburia 0.04664 0.04024 1.15900 0.24645
female Roseburia 2.05362 1.02396 2.00556 0.04490
time:intB Roseburia 0.11495 0.06153 1.86808 0.06175
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4971
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000003e8b9538>
Link: poisson
Intraclass Correlation (ICC): 0.5818463
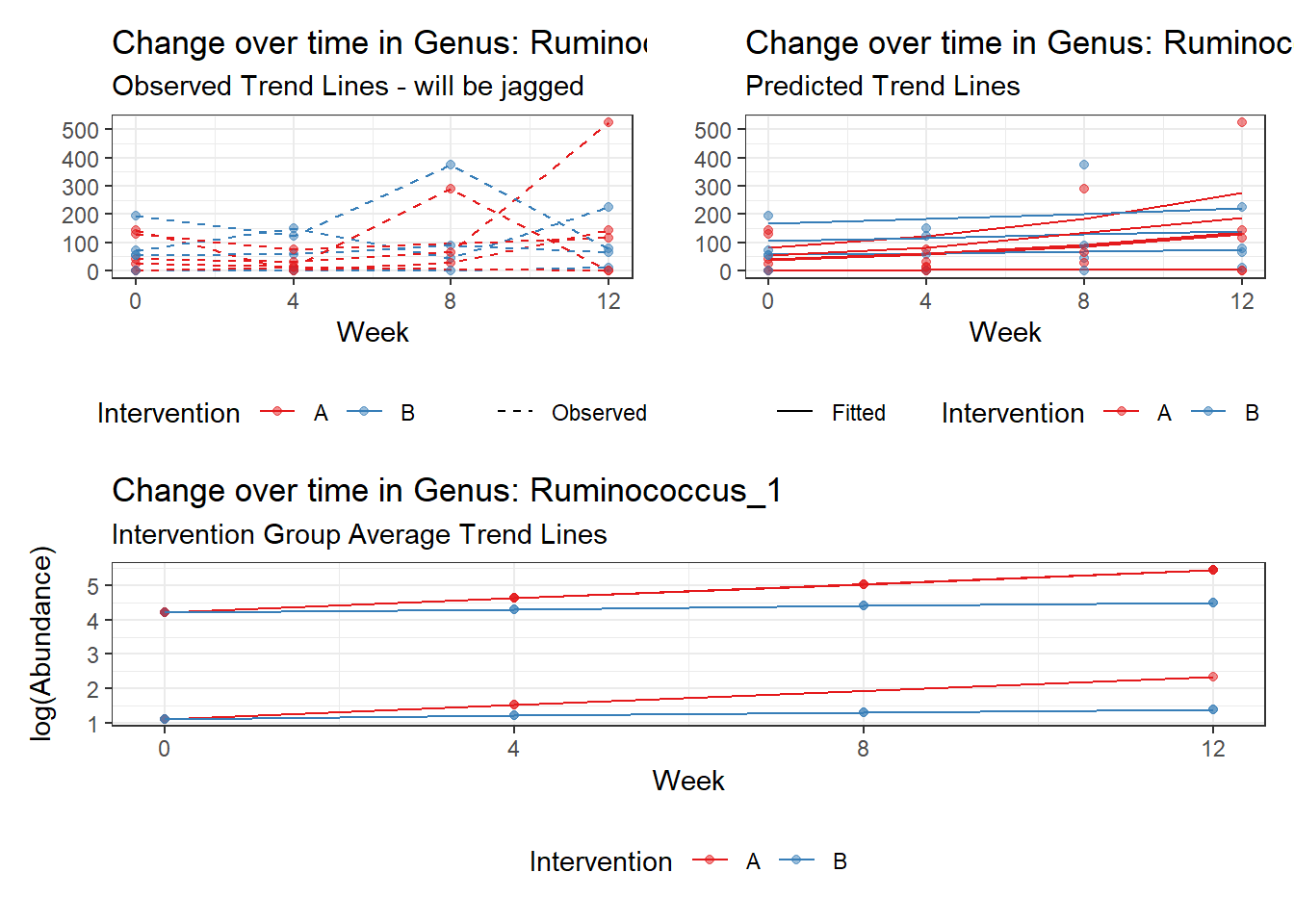
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 1.12590 0.63756 1.76595 0.07740
time Ruminococcus_1 0.40956 0.02274 18.01415 0.00000
female Ruminococcus_1 3.08815 0.77806 3.96904 0.00007
time:intB Ruminococcus_1 -0.31630 0.03223 -9.81479 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.1796
Significance test not available for random effectsgenus.fit7b <- glmm_microbiome(mydata=microbiome_data, model.number=7,
taxa.level="Genus", taxa.subset = backNames,
link="negbinom",
model="1 + time + intB:time + female + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x00000000350309c8>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 7.942102e-12
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 5.24356 0.95492 5.49110 0.00000
time Akkermansia -0.14371 0.44338 -0.32411 0.74585
female Akkermansia 0.71113 0.97493 0.72942 0.46575
time:intB Akkermansia 0.08040 0.50530 0.15912 0.87358
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.8182e-06
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x00000000350309c8>
Link: negbinomError in factory(refitNB, types = c("message", "warning"))(lastfit, theta = exp(t), : Downdated VtV is not positive definite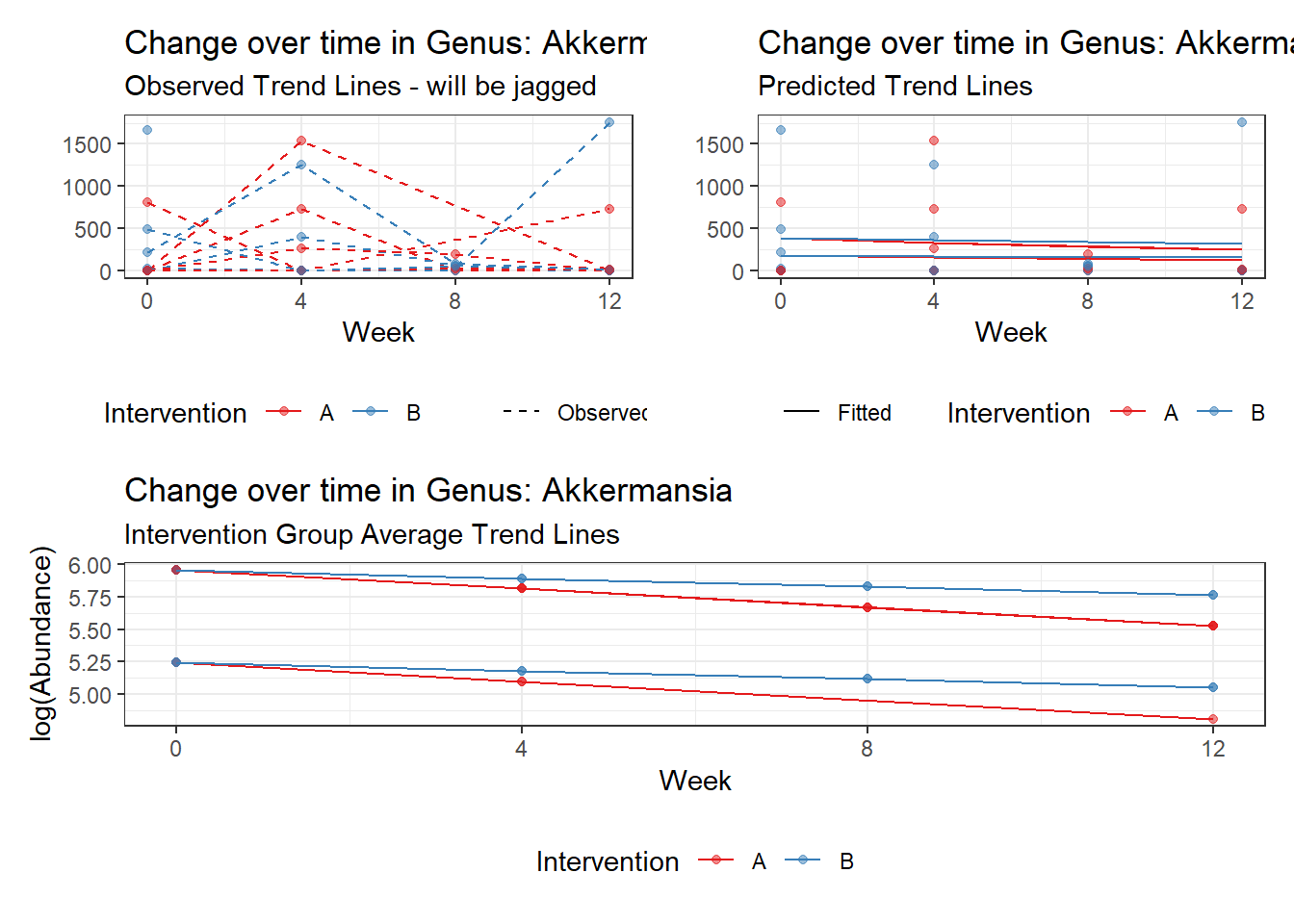
Covariate(s): Time and Ethnicity
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Ethnicity)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
genus.fit8 <- glmm_microbiome(mydata=microbiome_data, model.number=8,
taxa.level="Genus", taxa.subset = backNames,
link="poisson",
model="1 + time + intB:time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.8527204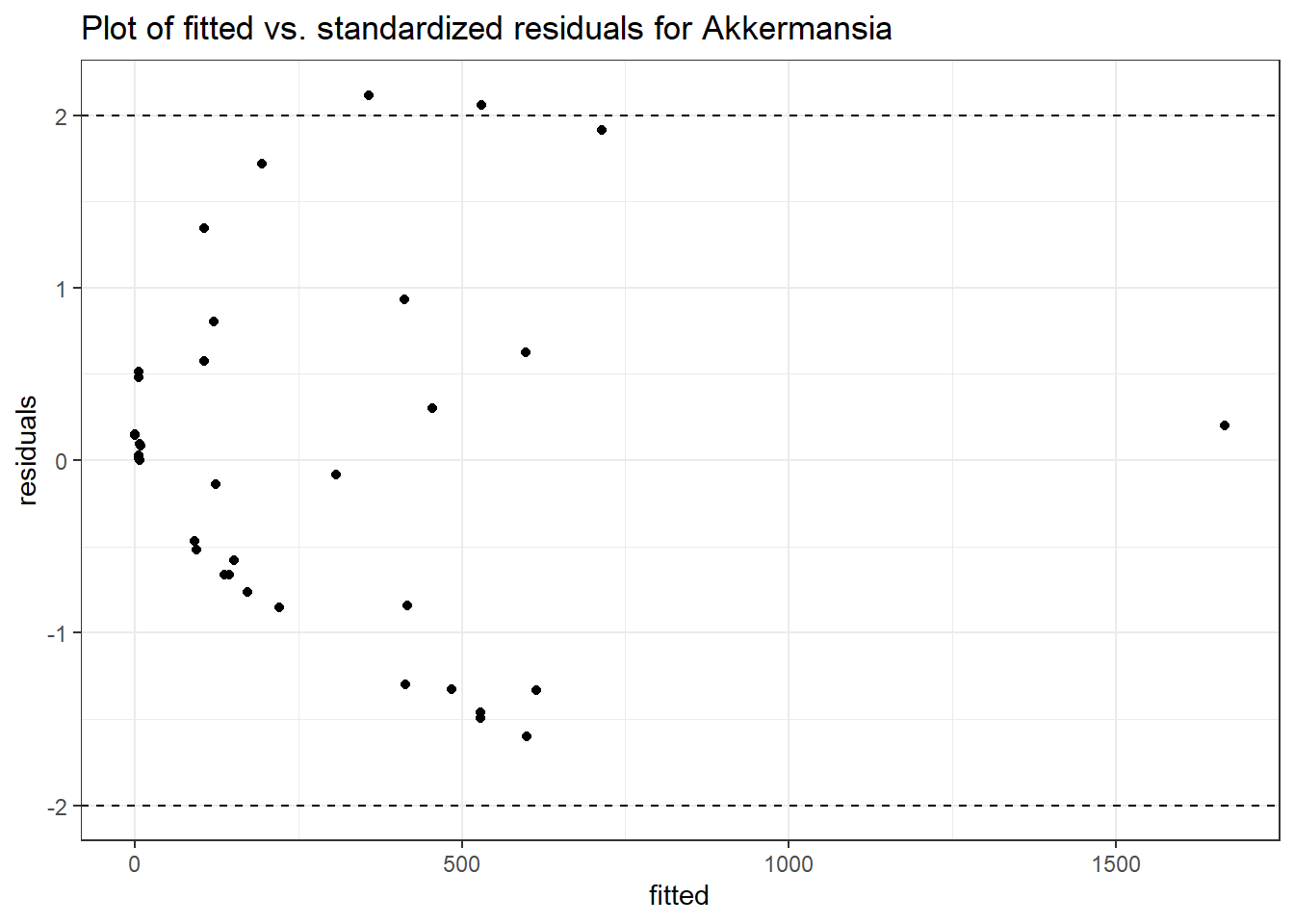
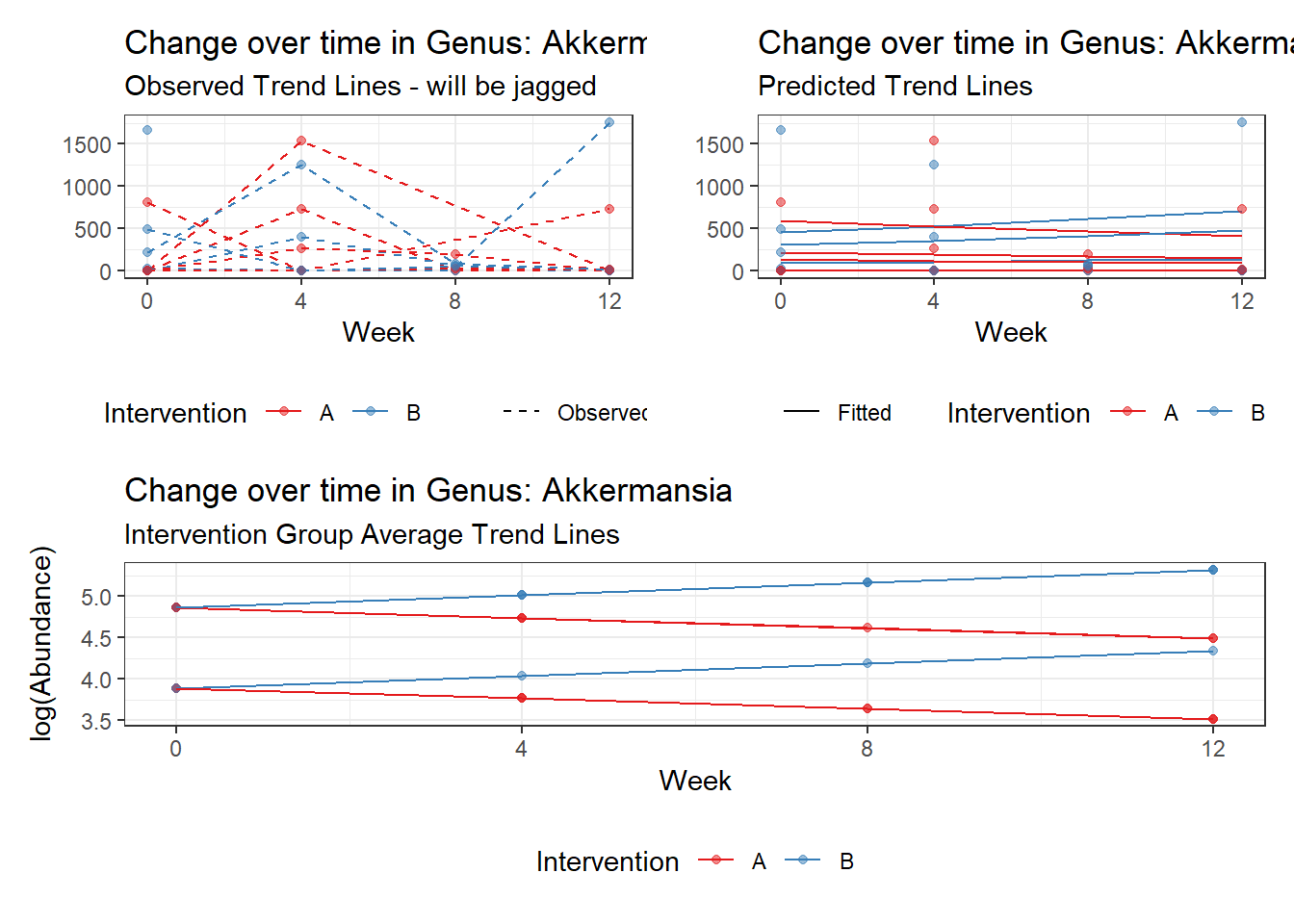
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 3.89165 1.20479 3.23014 0.00124
time Akkermansia -0.12314 0.01296 -9.49942 0.00000
hispanic Akkermansia 0.97273 1.51647 0.64145 0.52123
time:intB Akkermansia 0.27345 0.01882 14.52637 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4062
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.234196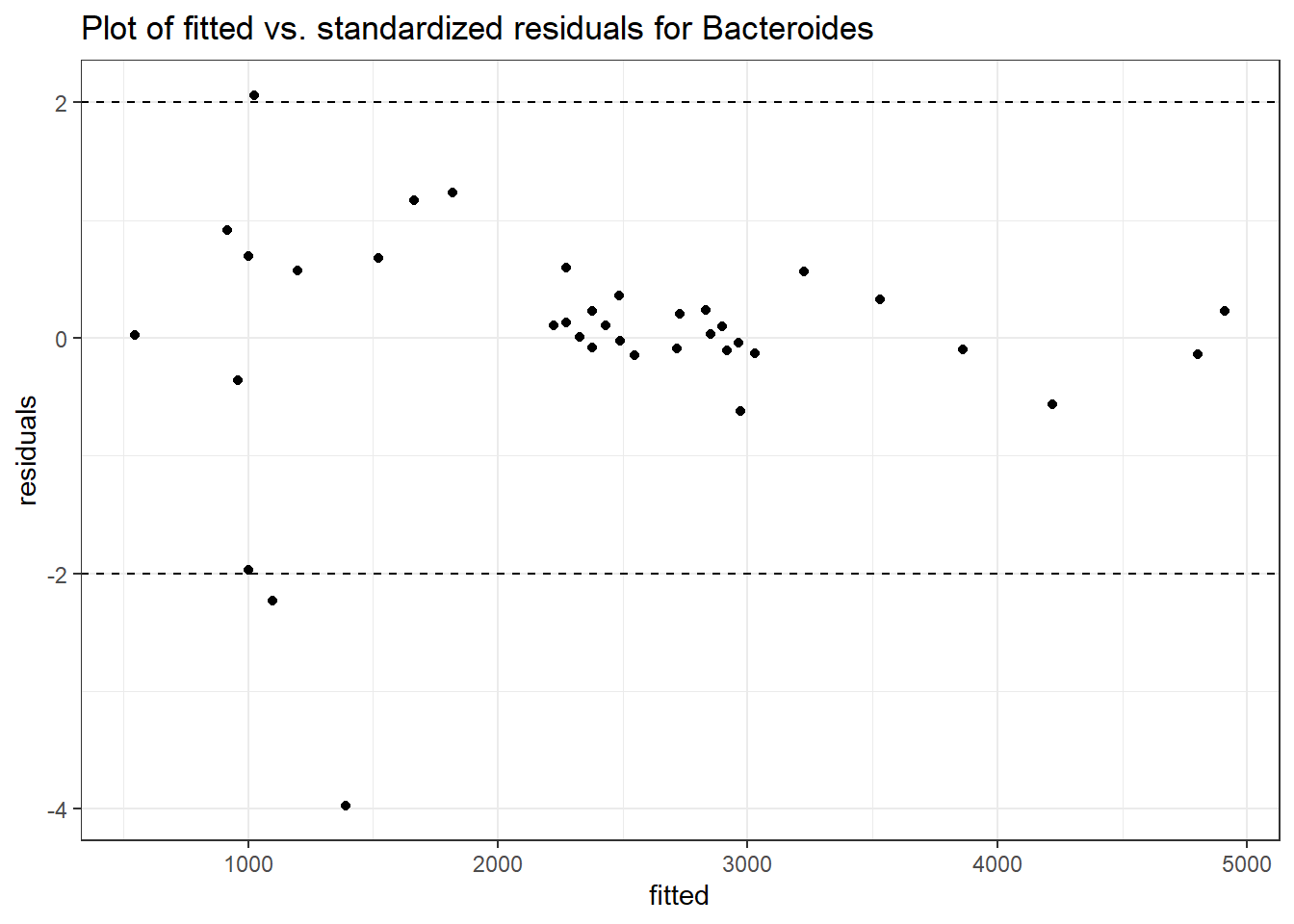

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 8.04618 0.27659 29.09097 0.00000
time Bacteroides -0.02257 0.00413 -5.46082 0.00000
hispanic Bacteroides -0.60323 0.34675 -1.73968 0.08192
time:intB Bacteroides -0.06698 0.00629 -10.65688 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.55301
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.53848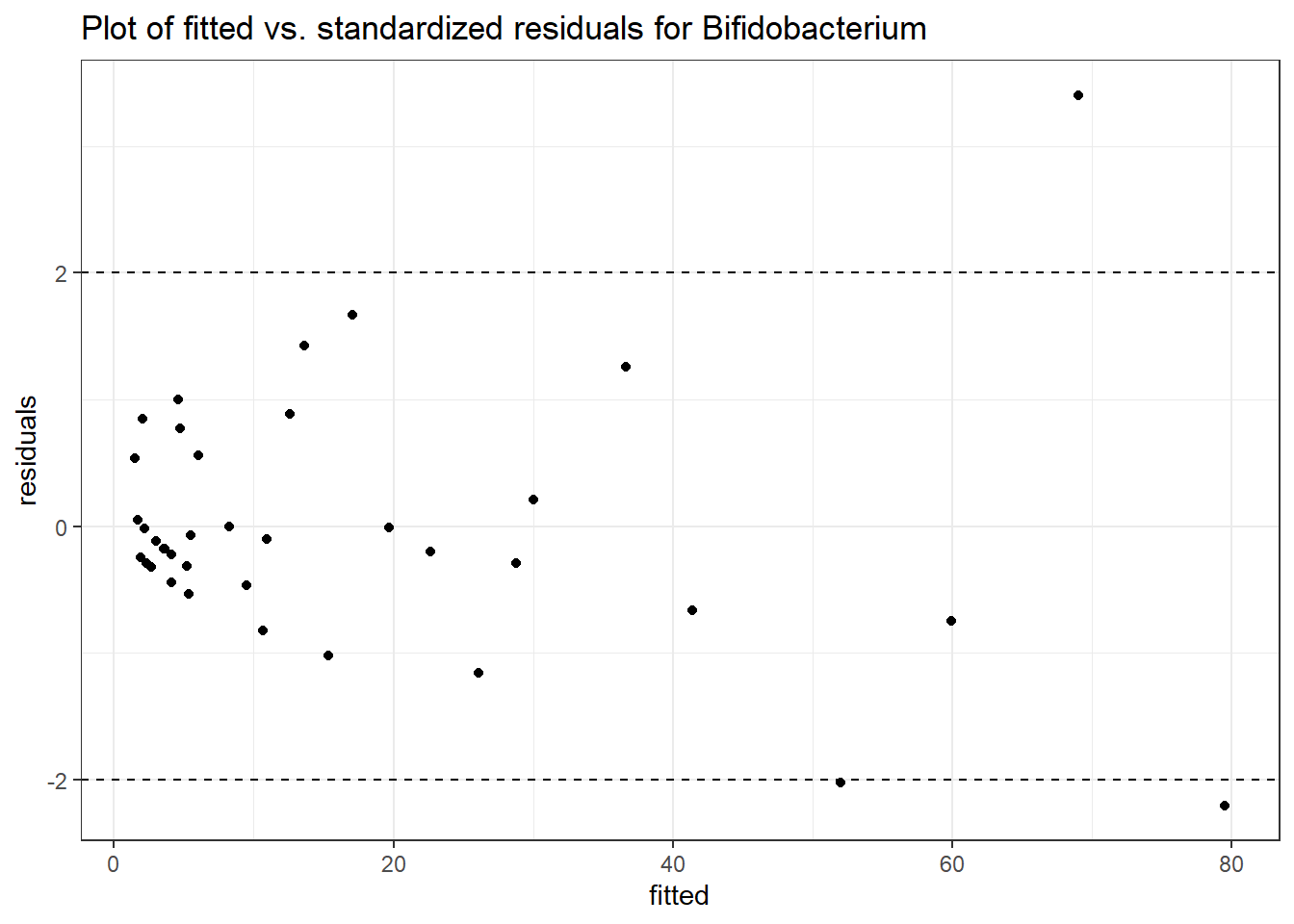

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.25182 0.55633 4.04766 0.00005
time Bifidobacterium -0.12129 0.06179 -1.96310 0.04963
hispanic Bifidobacterium 0.04729 0.69254 0.06828 0.94556
time:intB Bifidobacterium 0.26278 0.07587 3.46349 0.00053
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0802
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + intB:time + hispanic +
(1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.5717604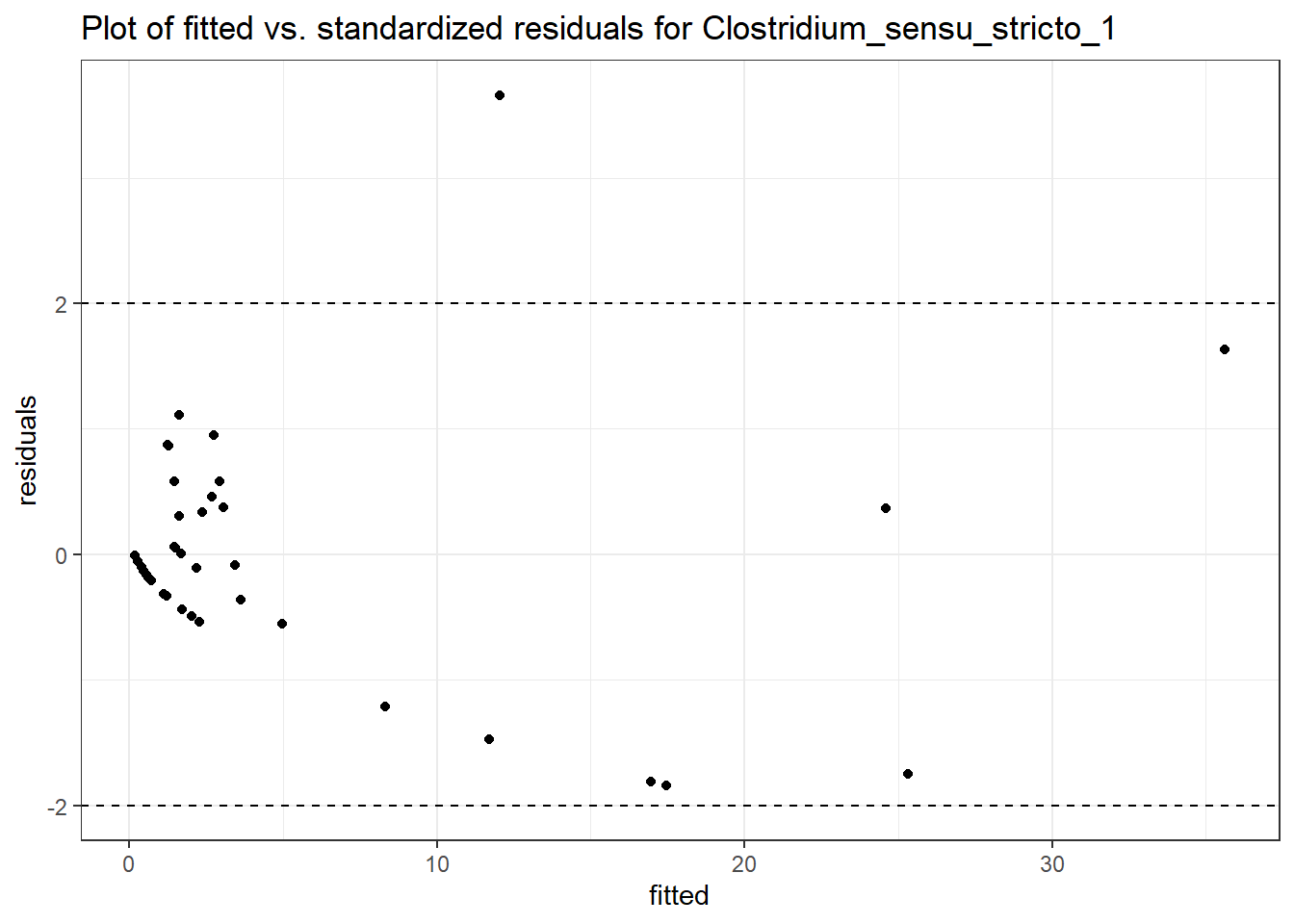

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.15649 0.65278 0.23973 0.81054
time Clostridium_sensu_stricto_1 -0.29923 0.14829 -2.01792 0.04360
hispanic Clostridium_sensu_stricto_1 0.82567 0.79309 1.04108 0.29784
time:intB Clostridium_sensu_stricto_1 0.67006 0.16318 4.10634 0.00004
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.1555
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.2647366

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.06227 0.30989 9.88171 0.00000
time Dorea 0.10111 0.04177 2.42047 0.01550
hispanic Dorea 0.39136 0.38476 1.01714 0.30909
time:intB Dorea -0.51266 0.05835 -8.78588 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.60005
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.2803461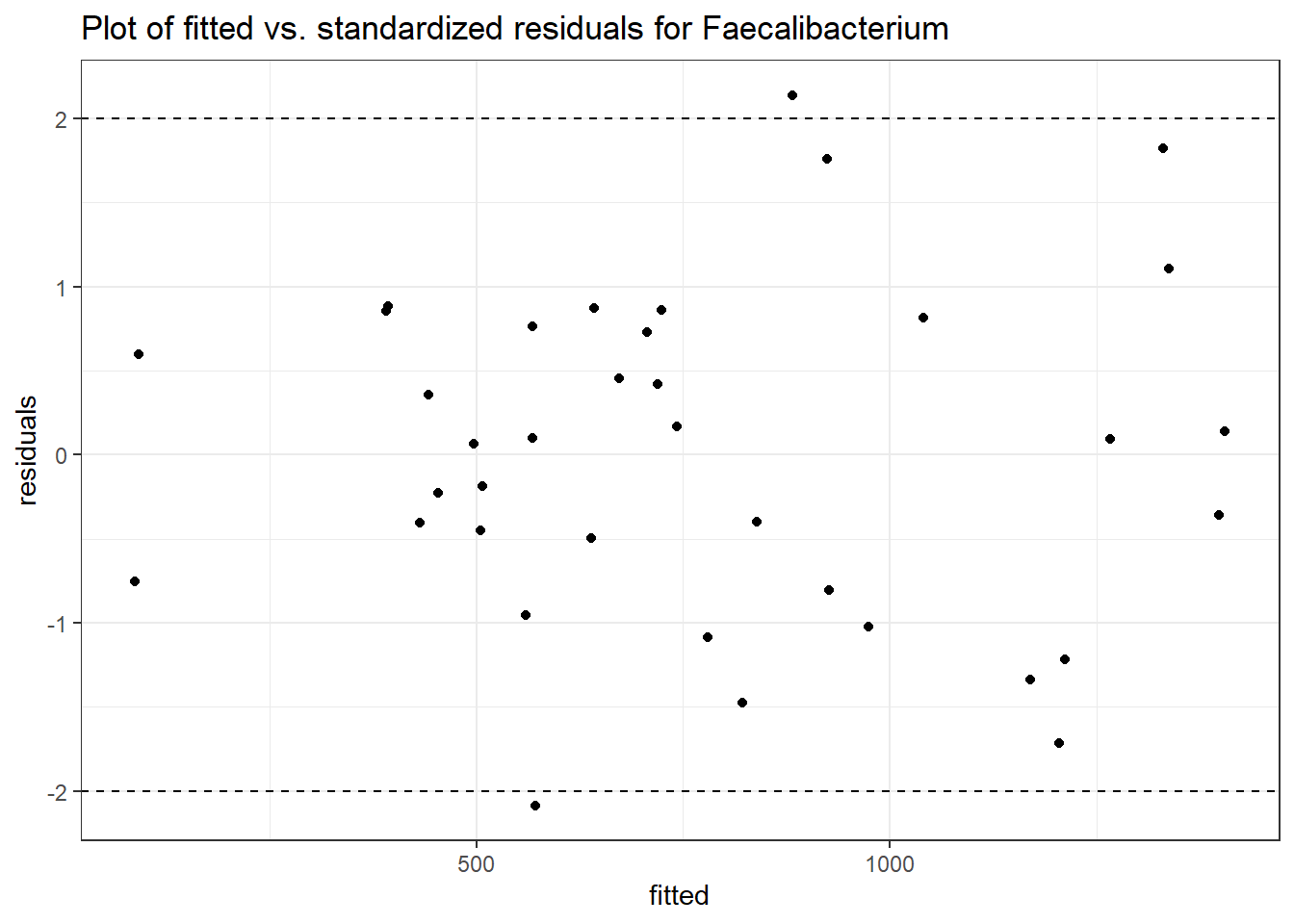
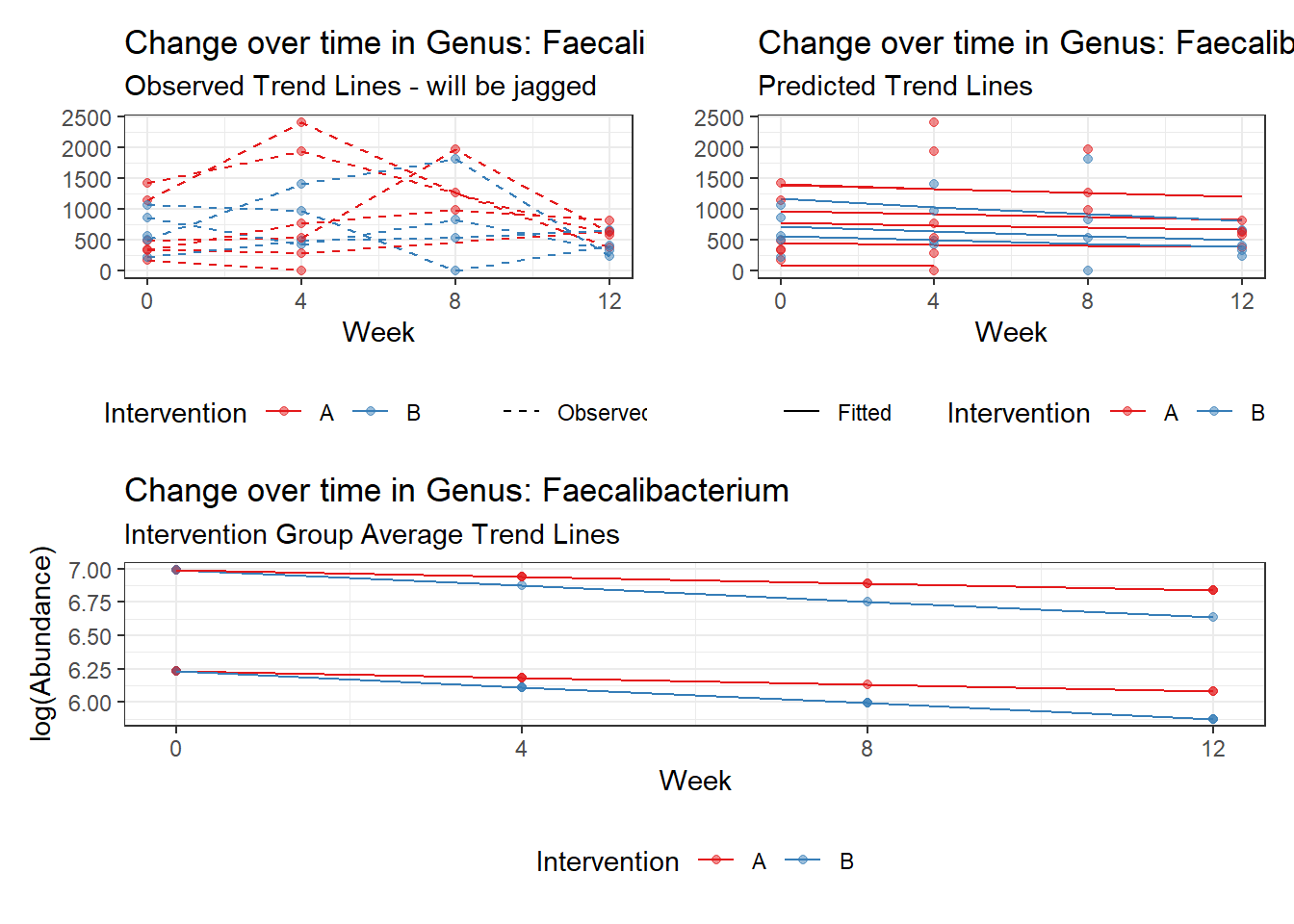
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.98881 0.31228 22.37991 0.00000
time Faecalibacterium -0.04946 0.00665 -7.43703 0.00000
hispanic Faecalibacterium -0.75943 0.39158 -1.93940 0.05245
time:intB Faecalibacterium -0.06839 0.01093 -6.25466 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.62414
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.3060503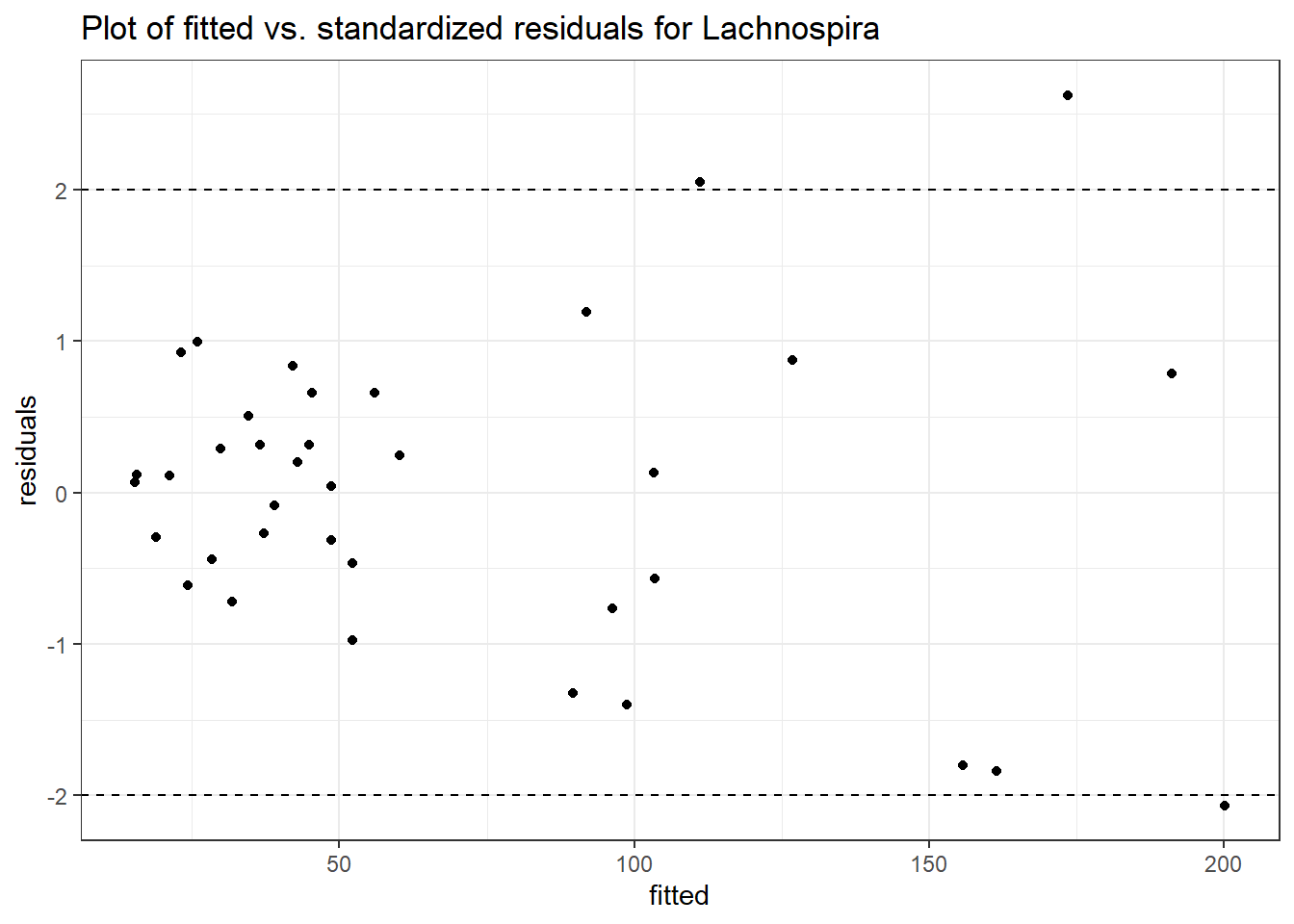

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.33533 0.33453 12.95932 0.00000
time Lachnospira 0.07154 0.02181 3.27962 0.00104
hispanic Lachnospira -0.87311 0.42015 -2.07810 0.03770
time:intB Lachnospira 0.13351 0.03690 3.61813 0.00030
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.6641
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.875729

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus -0.58524 1.70135 -0.34398 0.73086
time Lactobacillus -0.98369 0.09737 -10.10236 0.00000
hispanic Lactobacillus -1.11084 1.94769 -0.57034 0.56845
time:intB Lactobacillus 0.89484 0.52541 1.70314 0.08854
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6546
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.7560331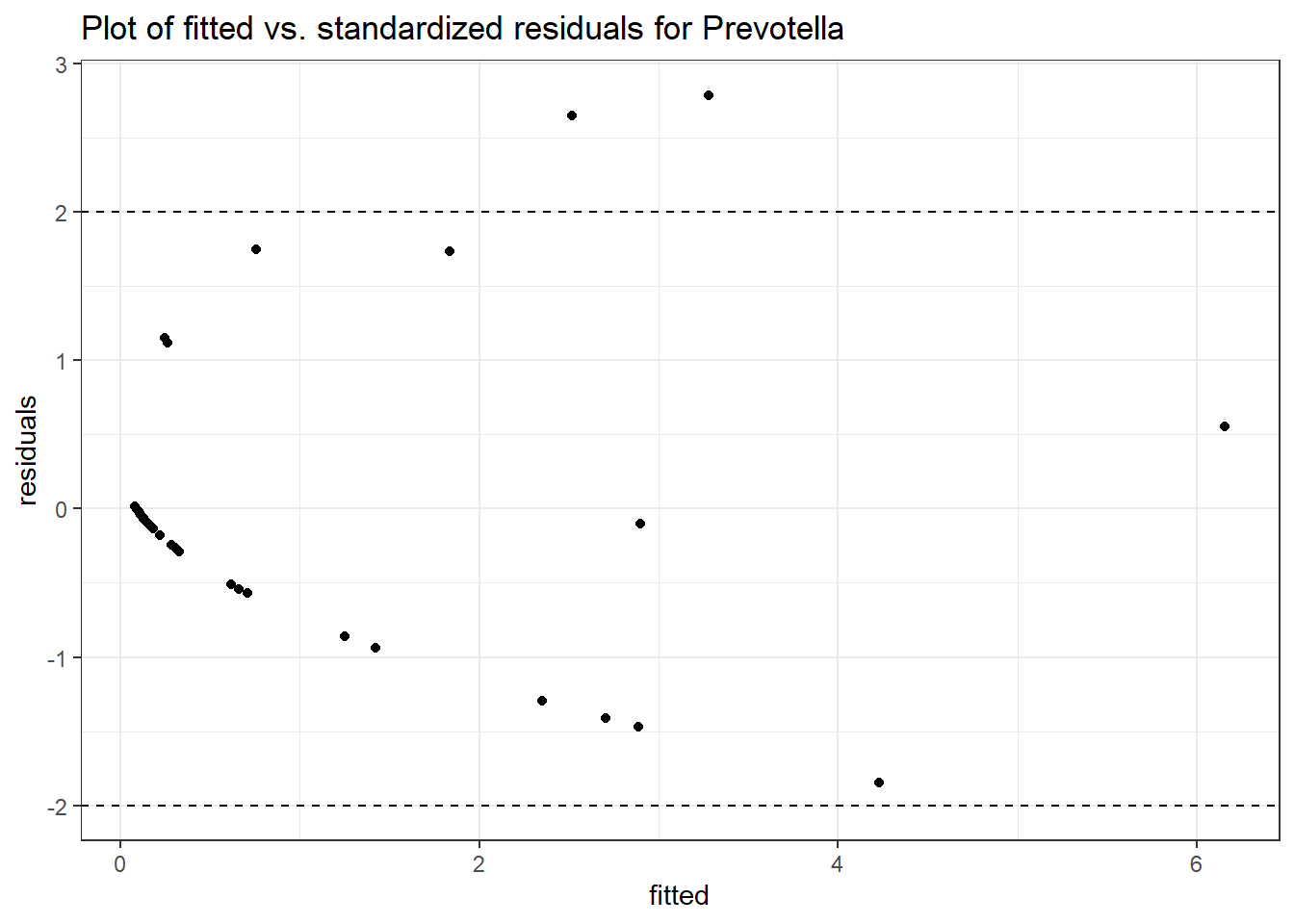
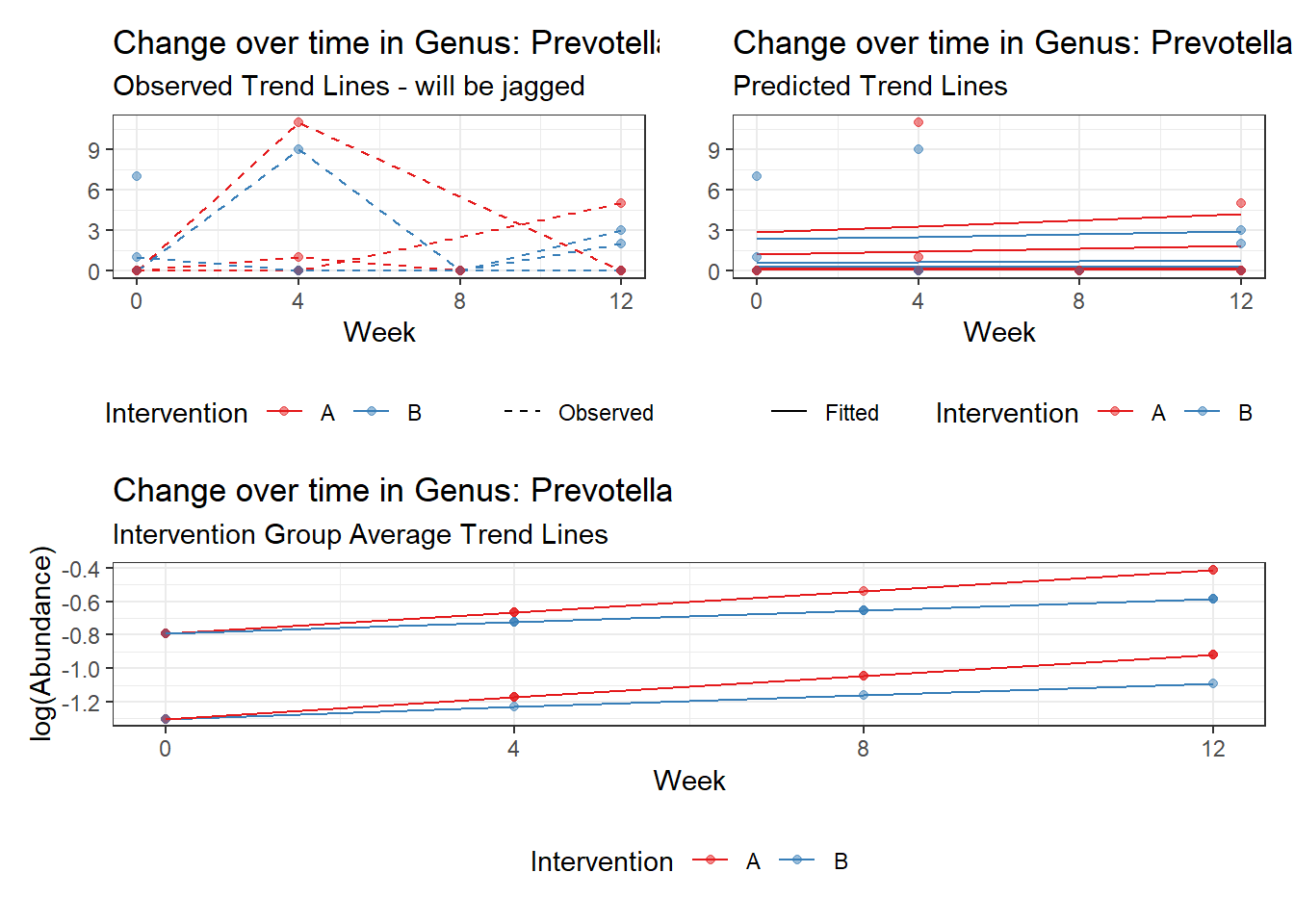
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.30094 1.05756 -1.23013 0.21865
time Prevotella 0.12711 0.18782 0.67680 0.49854
hispanic Prevotella 0.50714 1.27775 0.39690 0.69144
time:intB Prevotella -0.05692 0.28369 -0.20064 0.84098
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7604
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.7523701
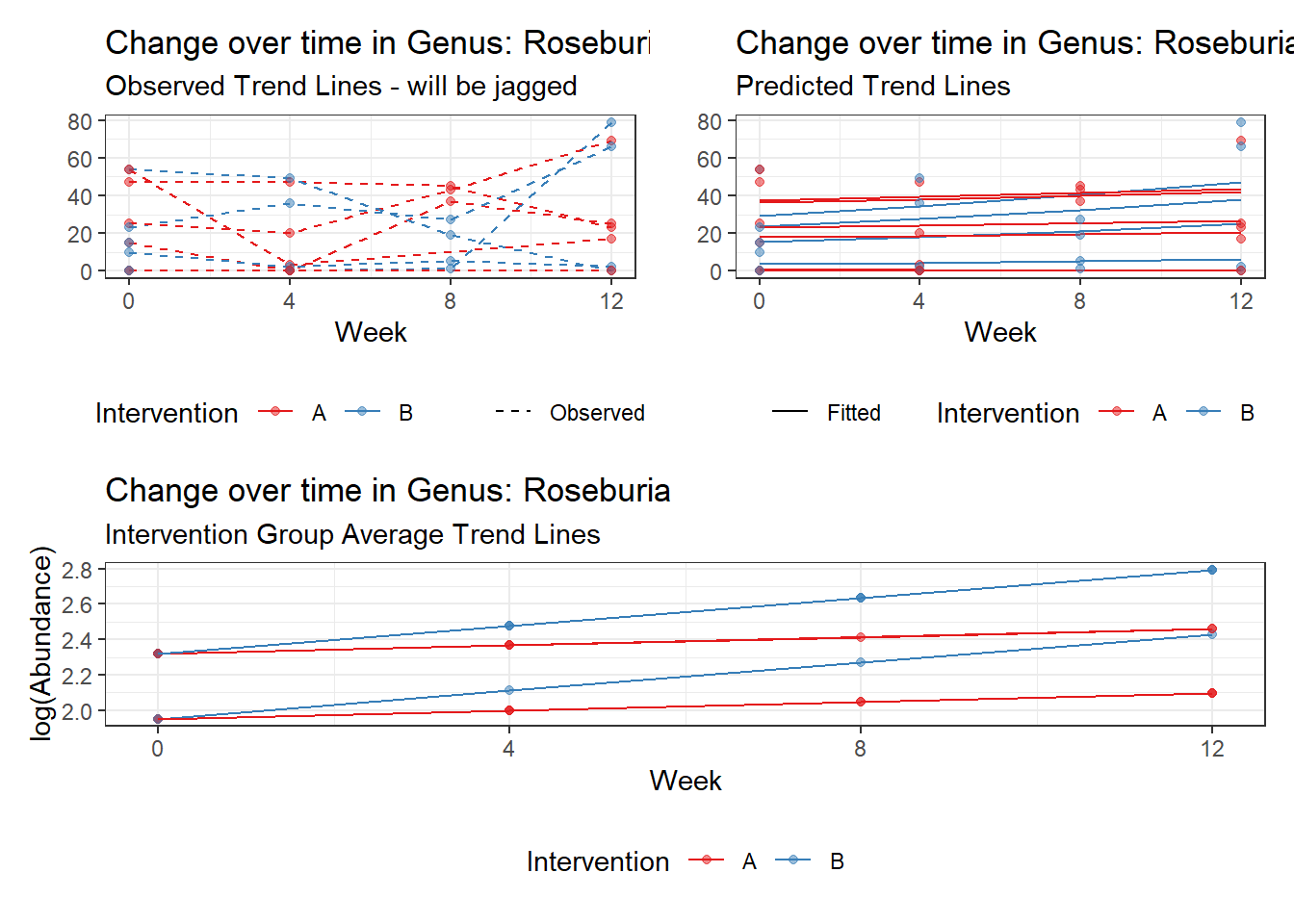
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 1.95104 0.90801 2.14870 0.03166
time Roseburia 0.04840 0.04029 1.20134 0.22962
hispanic Roseburia 0.36613 1.12403 0.32573 0.74463
time:intB Roseburia 0.11021 0.06152 1.79142 0.07323
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7431
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042cd4ff8>
Link: poisson
Intraclass Correlation (ICC): 0.7570993

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 2.15505 0.89040 2.42032 0.01551
time Ruminococcus_1 0.41167 0.02277 18.07694 0.00000
hispanic Ruminococcus_1 1.46818 1.11908 1.31195 0.18954
time:intB Ruminococcus_1 -0.32035 0.03222 -9.94124 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7655
Significance test not available for random effectsgenus.fit8b <- glmm_microbiome(mydata=microbiome_data, model.number=8,
taxa.level="Genus", taxa.subset = backNames,
link="negbinom",
model="1 + time + intB:time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Akkermansia ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 4.148378e-12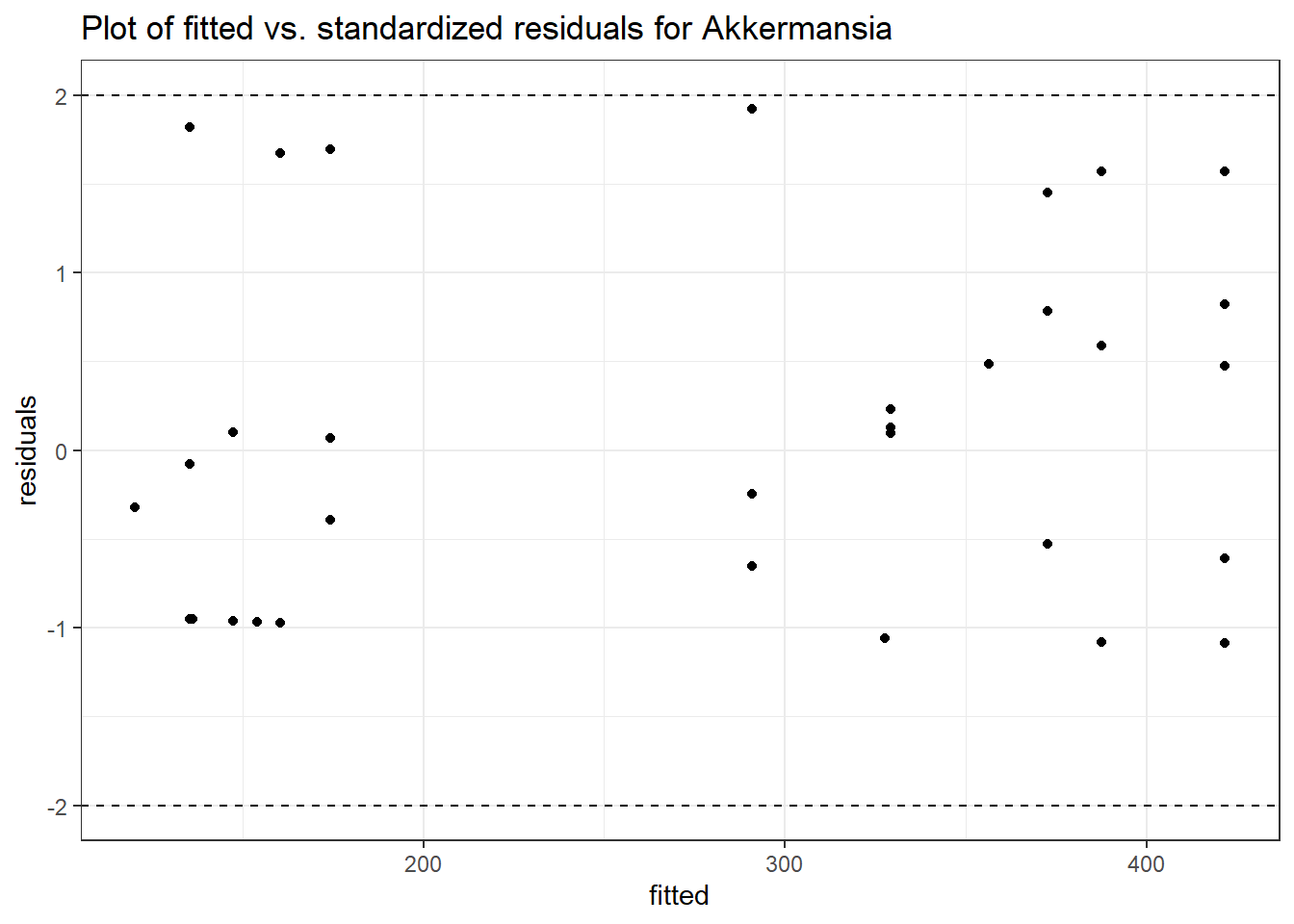

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Akkermansia 5.16109 0.93773 5.50382 0.00000
time Akkermansia -0.08415 0.47434 -0.17739 0.85920
hispanic Akkermansia 0.88331 1.06565 0.82889 0.40717
time:intB Akkermansia -0.03976 0.57503 -0.06914 0.94488
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.0368e-06
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroides ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinom
Intraclass Correlation (ICC): 0.1821143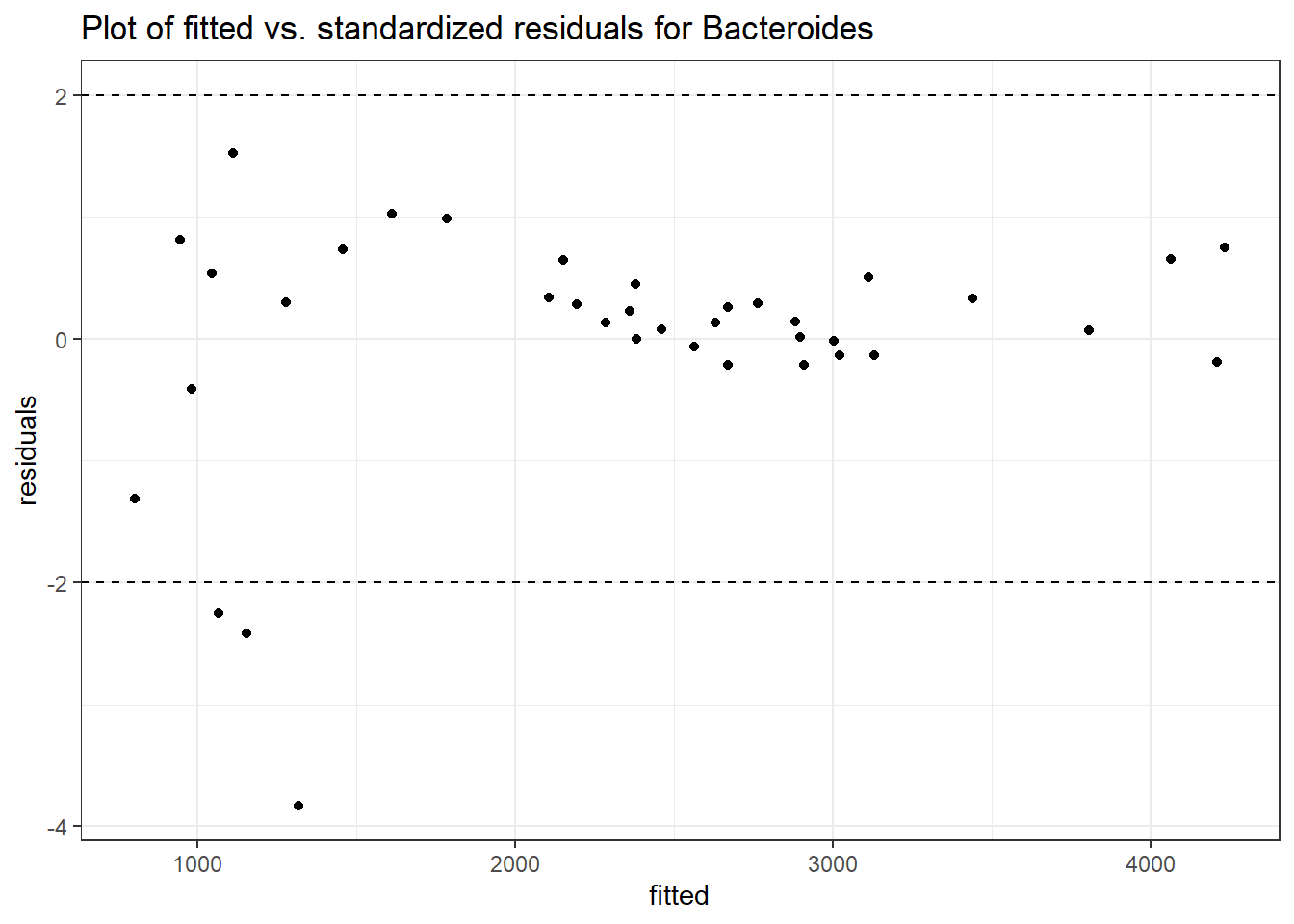
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroides 8.07384 0.25835 31.25163 0.00000
time Bacteroides -0.04133 0.05789 -0.71384 0.47533
hispanic Bacteroides -0.58246 0.31738 -1.83524 0.06647
time:intB Bacteroides -0.05932 0.08460 -0.70118 0.48319
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.47187
Significance test not available for random effects
#########################################
#########################################
Model: Bifidobacterium ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00471415 (tol = 0.002, component 1)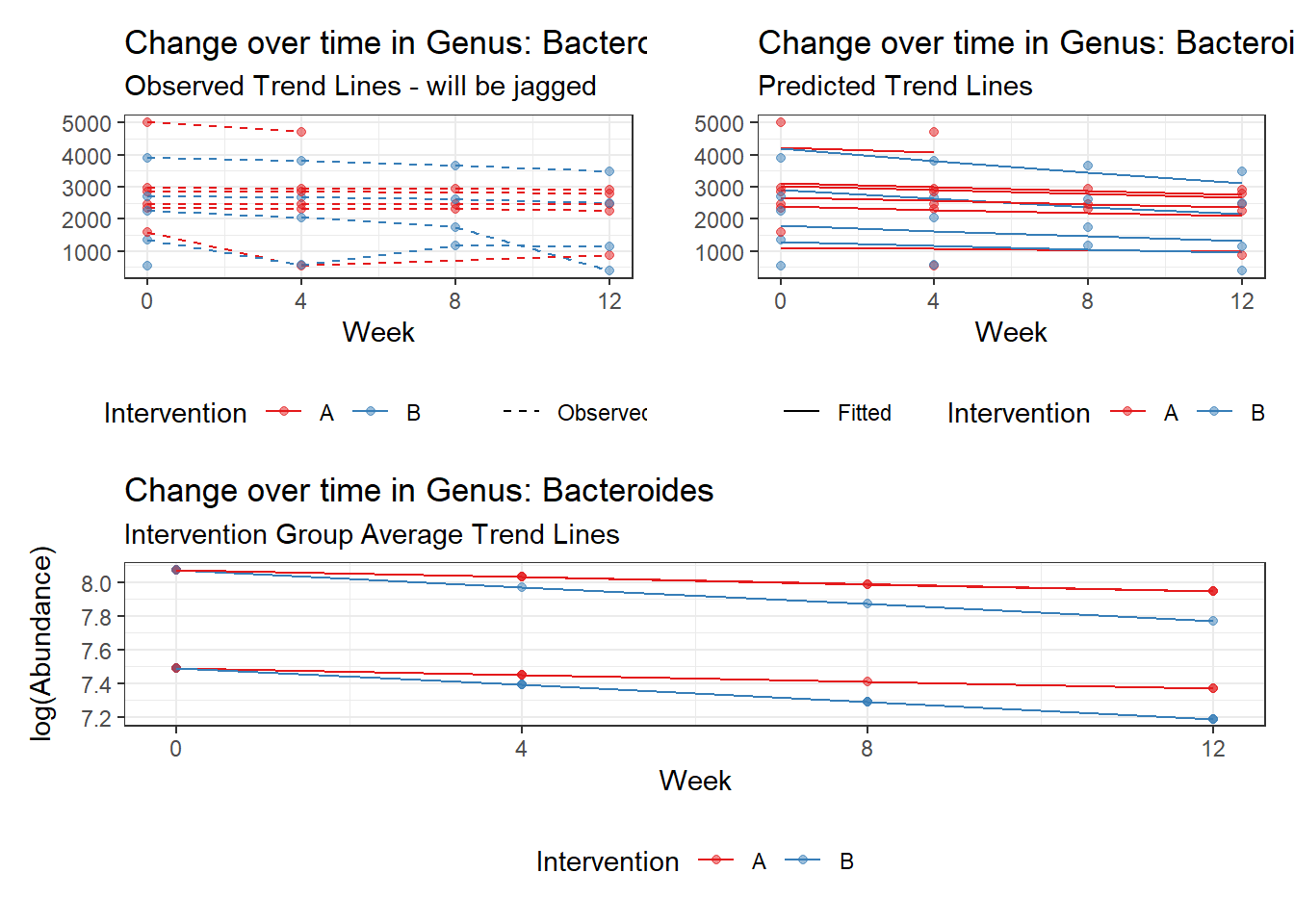
Intraclass Correlation (ICC): 0.2291176
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bifidobacterium 2.62399 0.72612 3.61369 0.00030
time Bifidobacterium -0.31188 0.31436 -0.99209 0.32115
hispanic Bifidobacterium -0.12457 0.67046 -0.18579 0.85261
time:intB Bifidobacterium 0.54982 0.36250 1.51674 0.12933
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.54517
Significance test not available for random effects
#########################################
#########################################
Model: Clostridium_sensu_stricto_1 ~ 1 + time + intB:time + hispanic +
(1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomboundary (singular) fit: see ?isSingular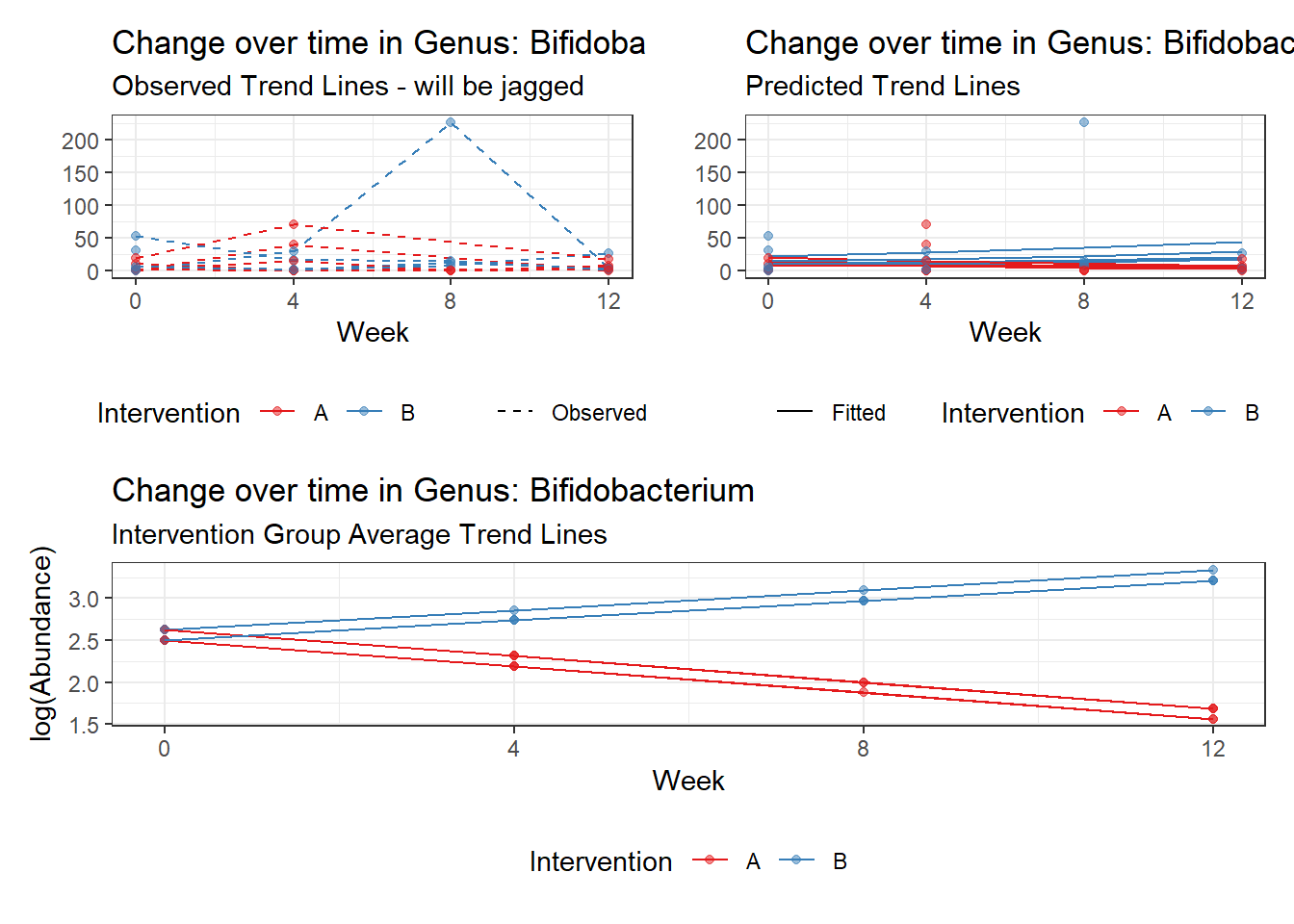
Intraclass Correlation (ICC): 1.014214e-11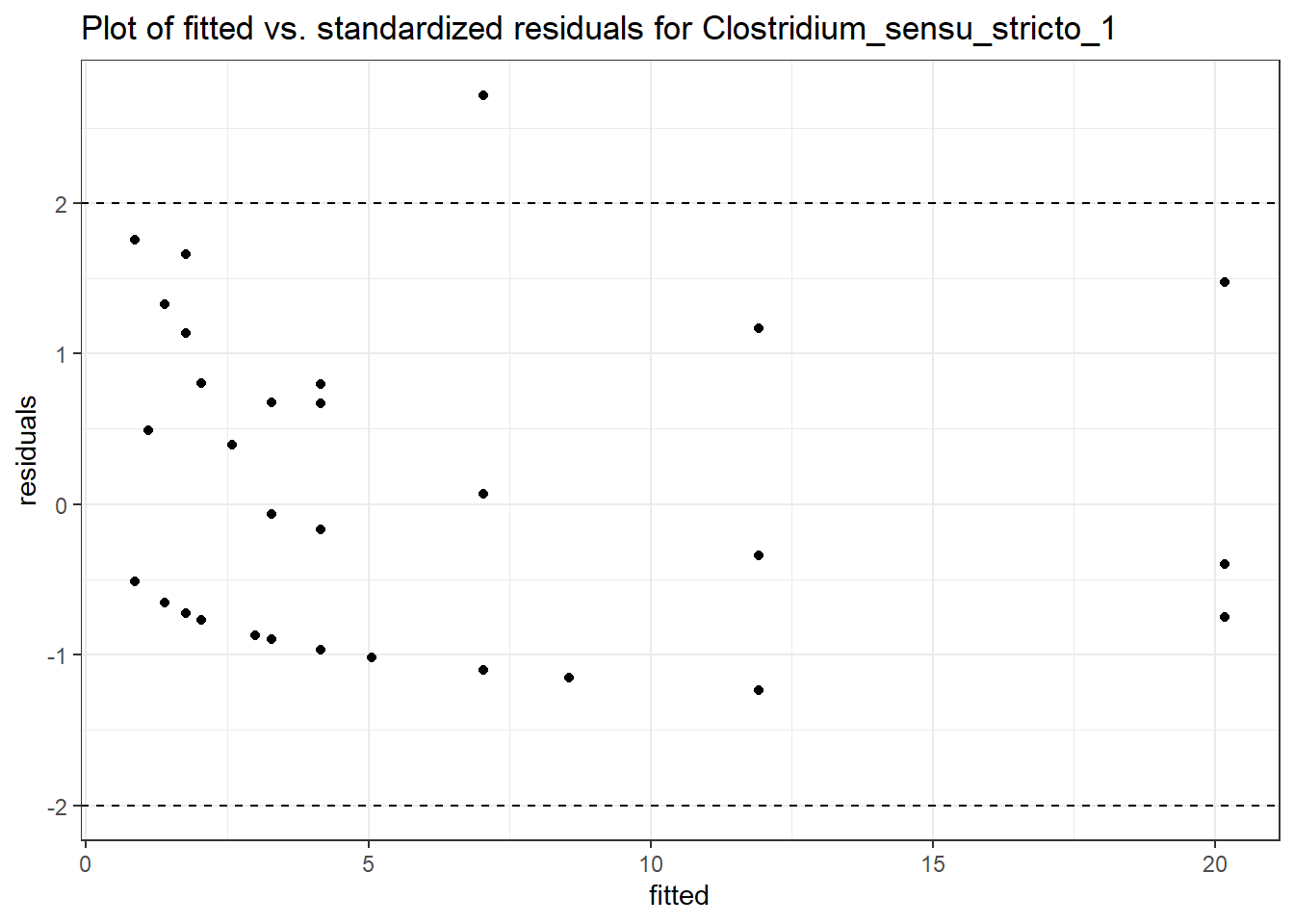
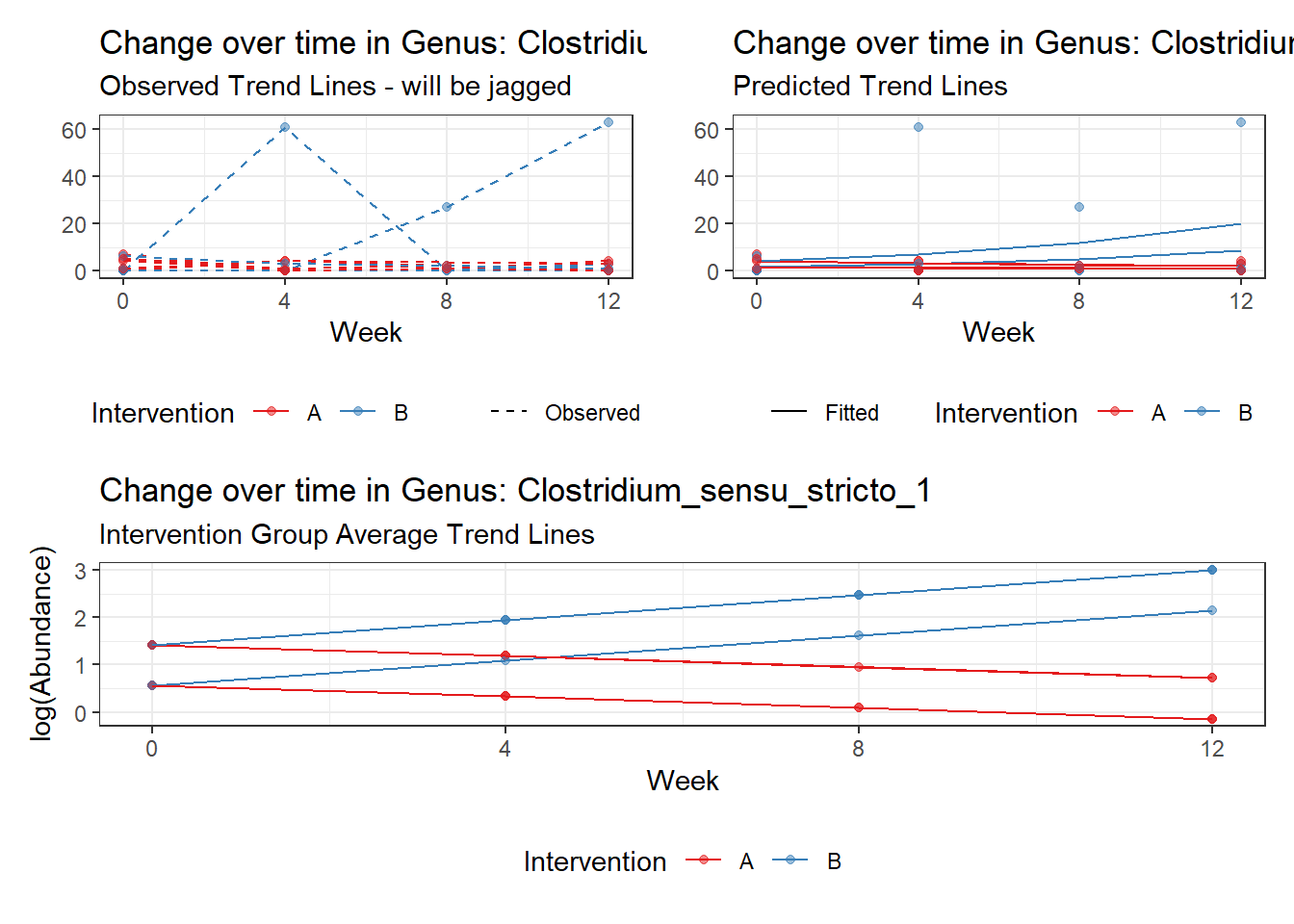
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Clostridium_sensu_stricto_1 0.56799 0.63134 0.89966 0.36830
time Clostridium_sensu_stricto_1 -0.23686 0.31931 -0.74179 0.45821
hispanic Clostridium_sensu_stricto_1 0.85691 0.75624 1.13312 0.25716
time:intB Clostridium_sensu_stricto_1 0.76336 0.41791 1.82661 0.06776
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.1847e-06
Significance test not available for random effects
#########################################
#########################################
Model: Dorea ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinom
Intraclass Correlation (ICC): 0.02269486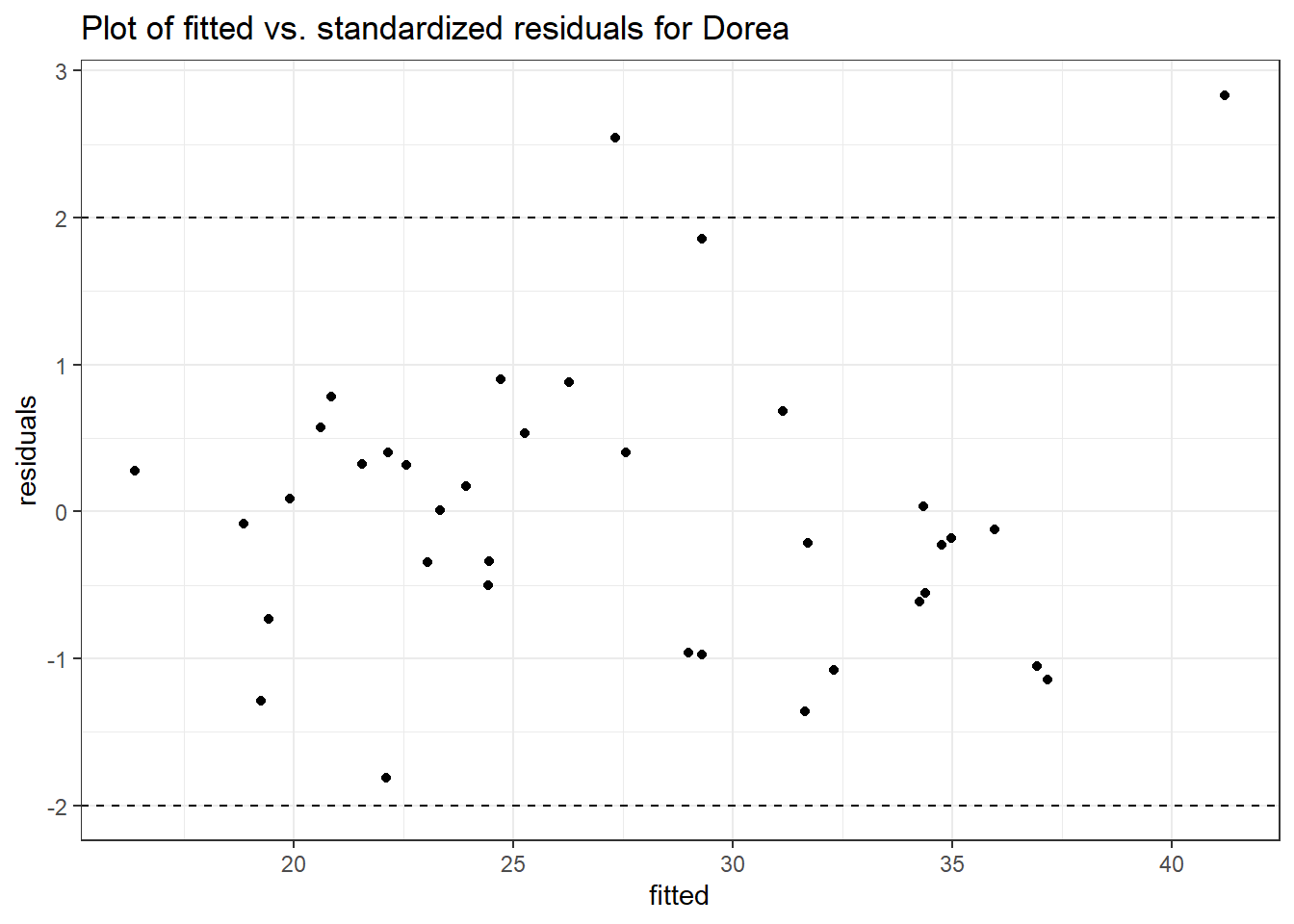
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Dorea 3.22667 0.24783 13.01960 0.00000
time Dorea -0.07950 0.12344 -0.64405 0.51954
hispanic Dorea 0.36613 0.26471 1.38314 0.16662
time:intB Dorea -0.09097 0.14435 -0.63018 0.52858
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.15239
Significance test not available for random effects
#########################################
#########################################
Model: Faecalibacterium ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomboundary (singular) fit: see ?isSingular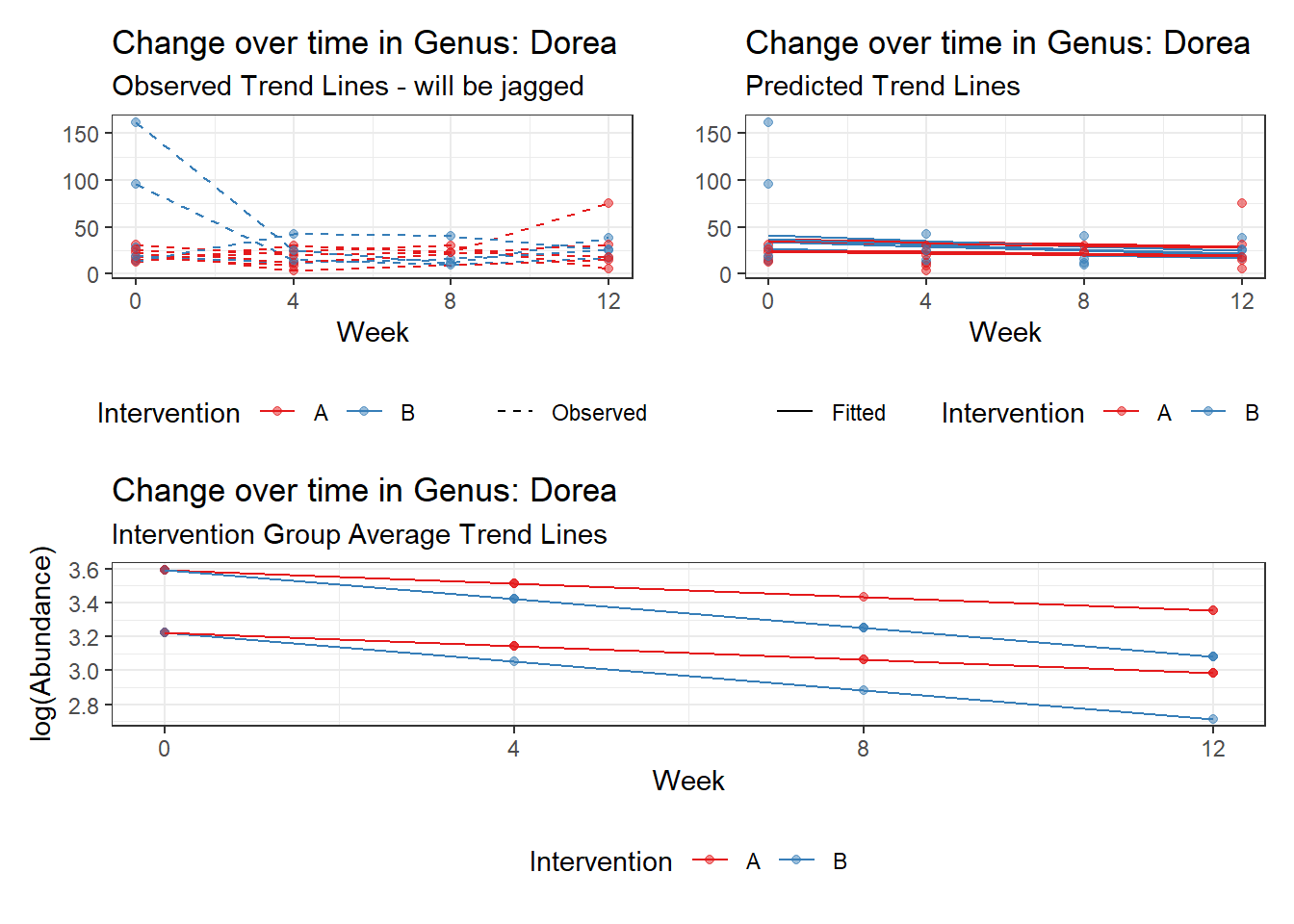
Intraclass Correlation (ICC): 1.142862e-10
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Faecalibacterium 6.90764 0.30638 22.54567 0.00000
time Faecalibacterium 0.01939 0.16941 0.11445 0.90888
hispanic Faecalibacterium -0.48581 0.34325 -1.41531 0.15698
time:intB Faecalibacterium -0.05584 0.19401 -0.28781 0.77349
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.069e-05
Significance test not available for random effects
#########################################
#########################################
Model: Lachnospira ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced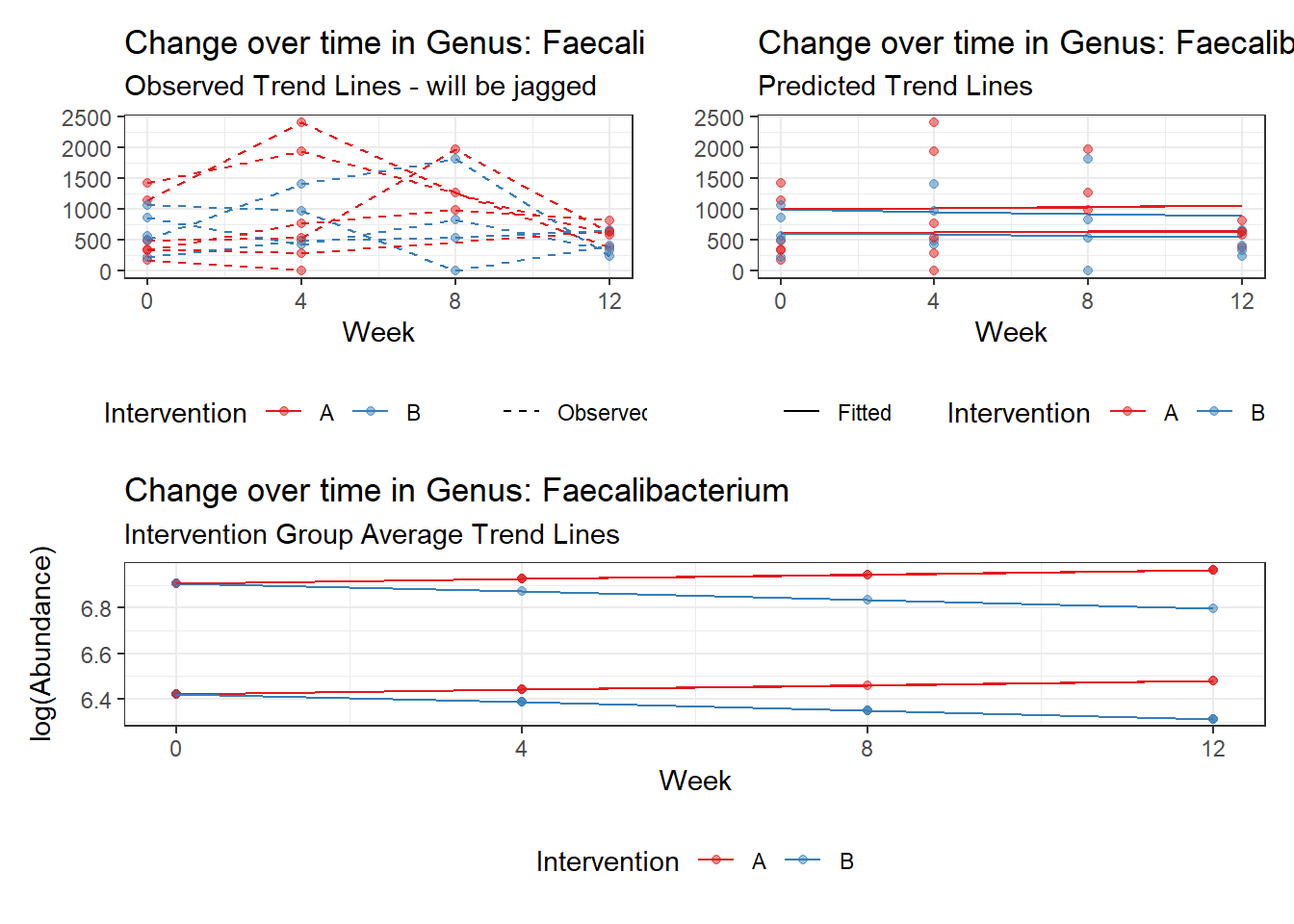
Intraclass Correlation (ICC): 0.3055965
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lachnospira 4.33432 0.33414 12.97159 0.00000
time Lachnospira 0.07298 0.02213 3.29800 0.00097
hispanic Lachnospira -0.87226 0.41966 -2.07848 0.03767
time:intB Lachnospira 0.13122 0.03726 3.52156 0.00043
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.66339
Significance test not available for random effects
#########################################
#########################################
Model: Lactobacillus ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomboundary (singular) fit: see ?isSingular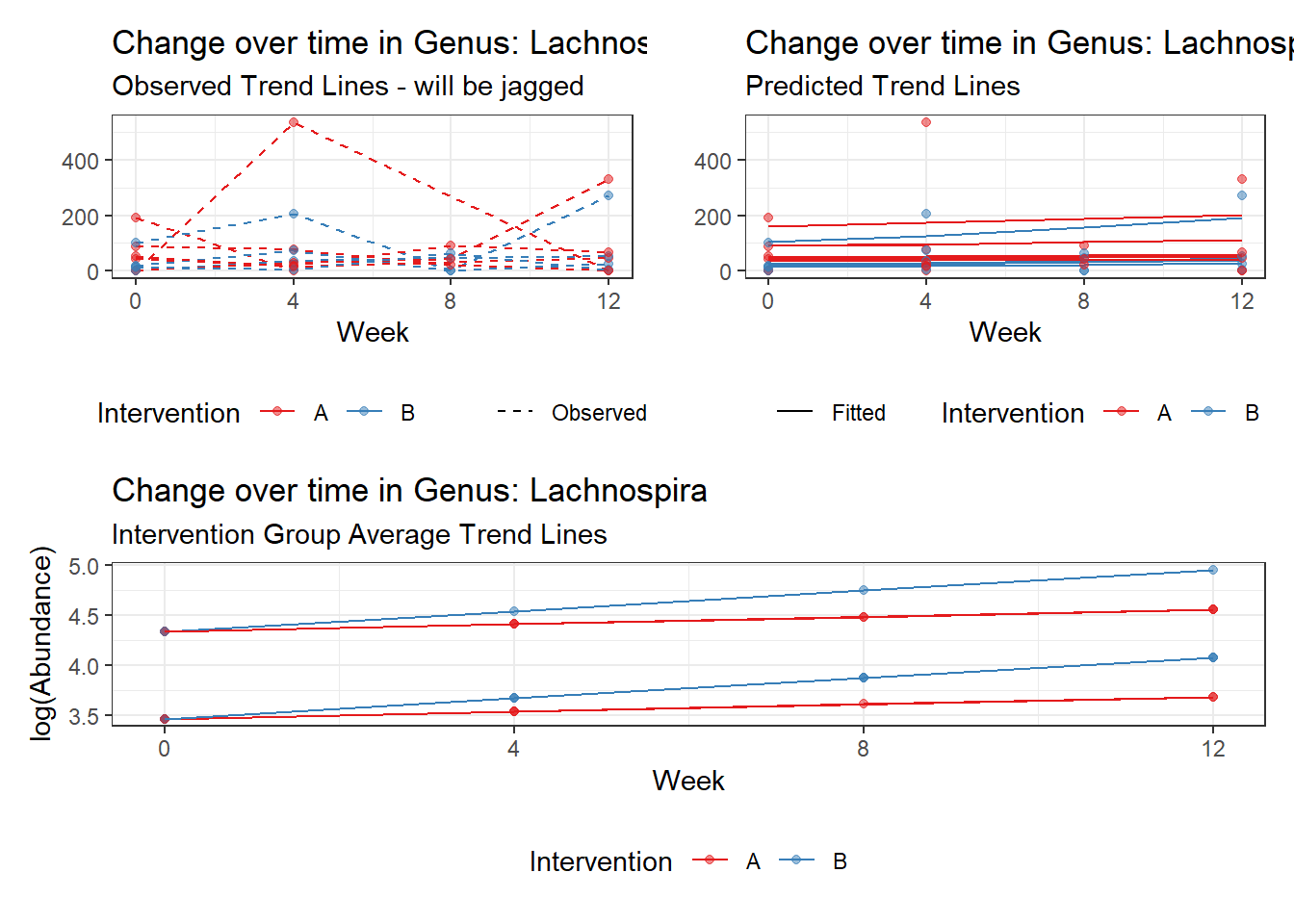
Intraclass Correlation (ICC): 3.366582e-13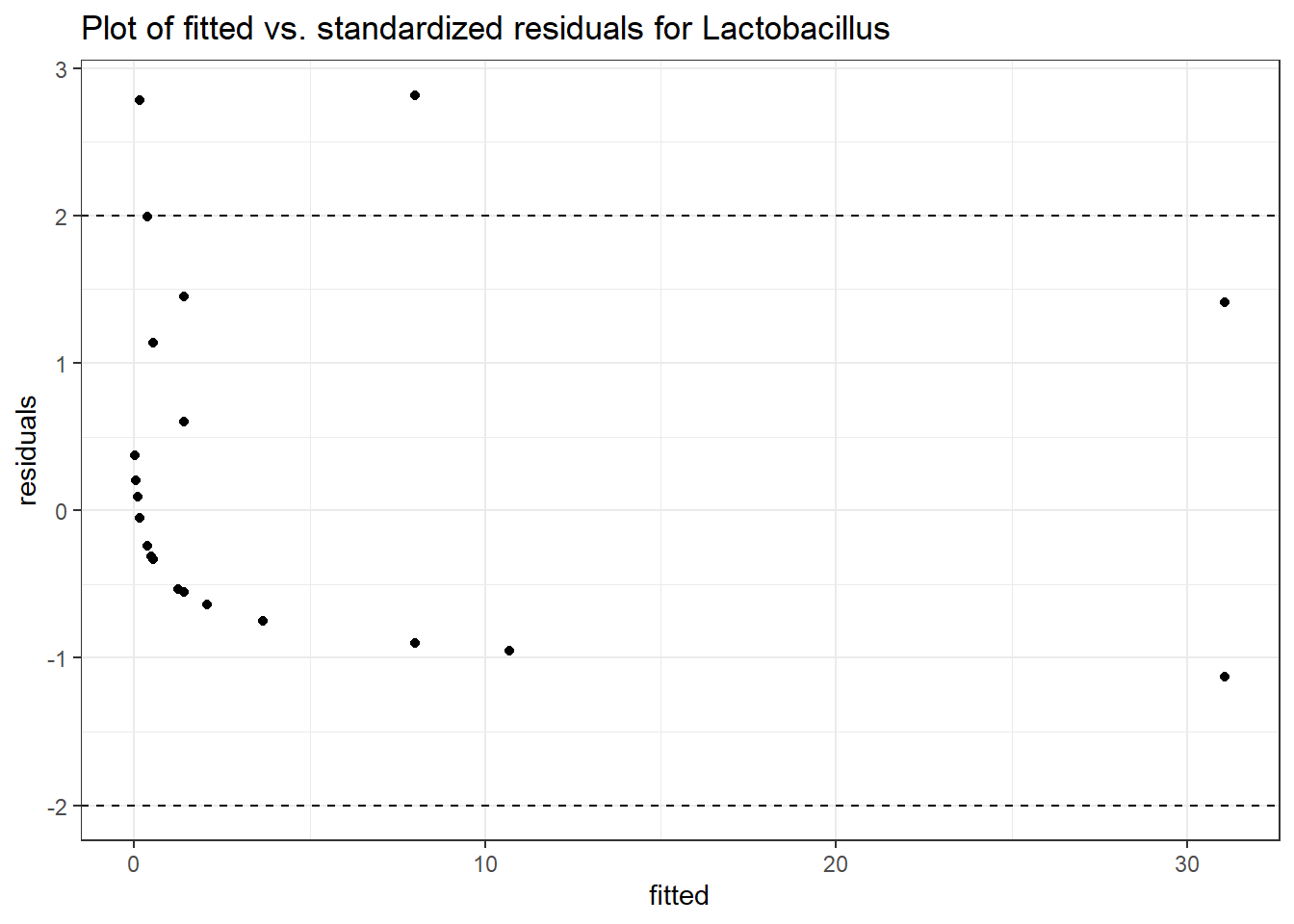
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lactobacillus 3.43660 1.34915 2.54724 0.01086
time Lactobacillus -1.35589 0.93013 -1.45774 0.14491
hispanic Lactobacillus -3.09202 1.54195 -2.00527 0.04493
time:intB Lactobacillus 0.28896 1.04561 0.27636 0.78227
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 5.8022e-07
Significance test not available for random effects
#########################################
#########################################
Model: Prevotella ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 2.1924e-12
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Prevotella -1.43347 1.76389 -0.81268 0.41640
time Prevotella 0.42632 0.74361 0.57331 0.56643
hispanic Prevotella 1.45803 1.52300 0.95734 0.33840
time:intB Prevotella -0.14075 0.60197 -0.23382 0.81513
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4807e-06
Significance test not available for random effects
#########################################
#########################################
Model: Roseburia ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0136036 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular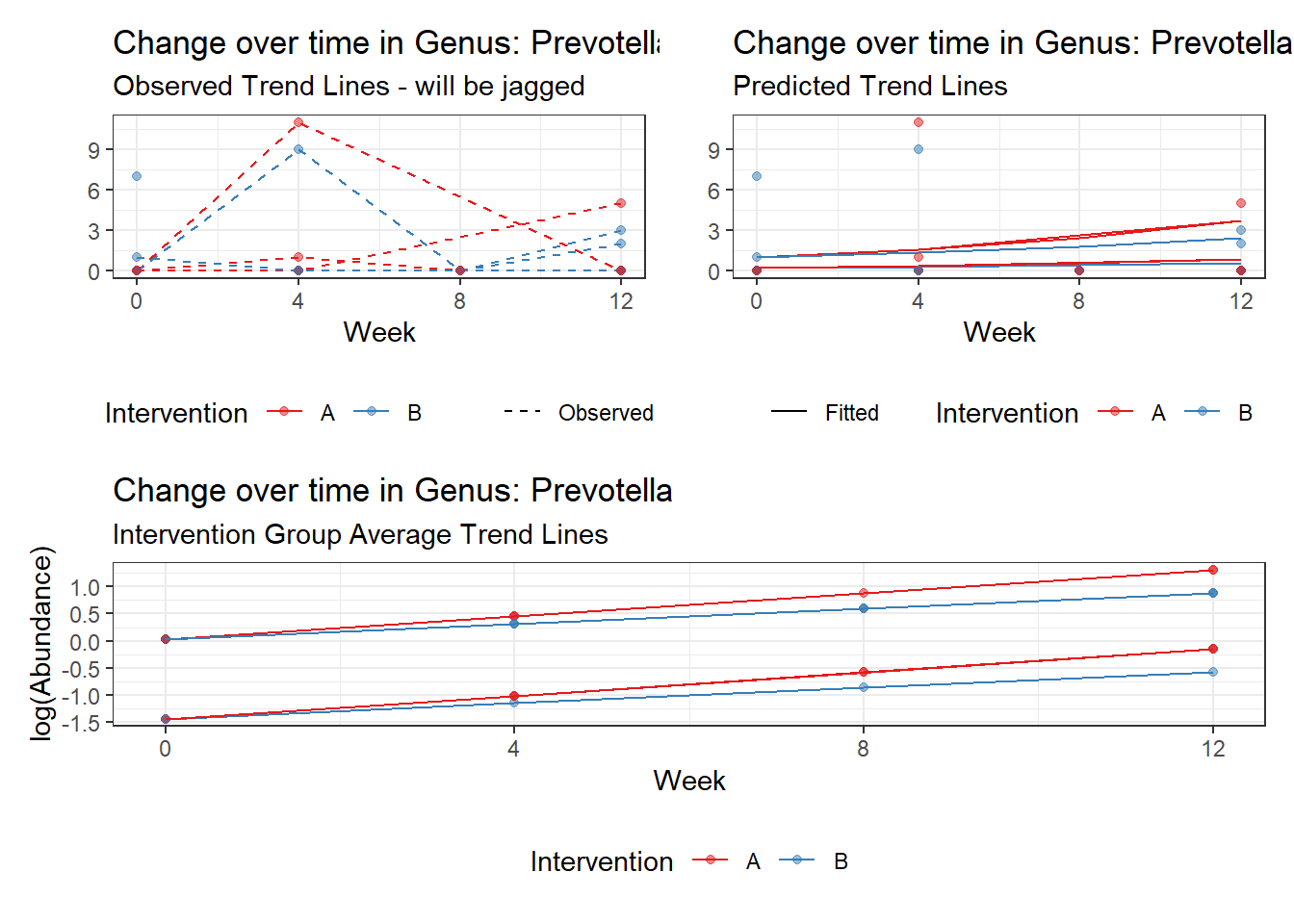
Intraclass Correlation (ICC): 4.798754e-12
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Roseburia 2.78430 0.52292 5.32451 0.00000
time Roseburia 0.16550 0.25247 0.65553 0.51212
hispanic Roseburia 0.21729 0.51639 0.42078 0.67391
time:intB Roseburia -0.02607 0.27146 -0.09602 0.92351
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1906e-06
Significance test not available for random effects
#########################################
#########################################
Model: Ruminococcus_1 ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x0000000042fe0538>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00629326 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular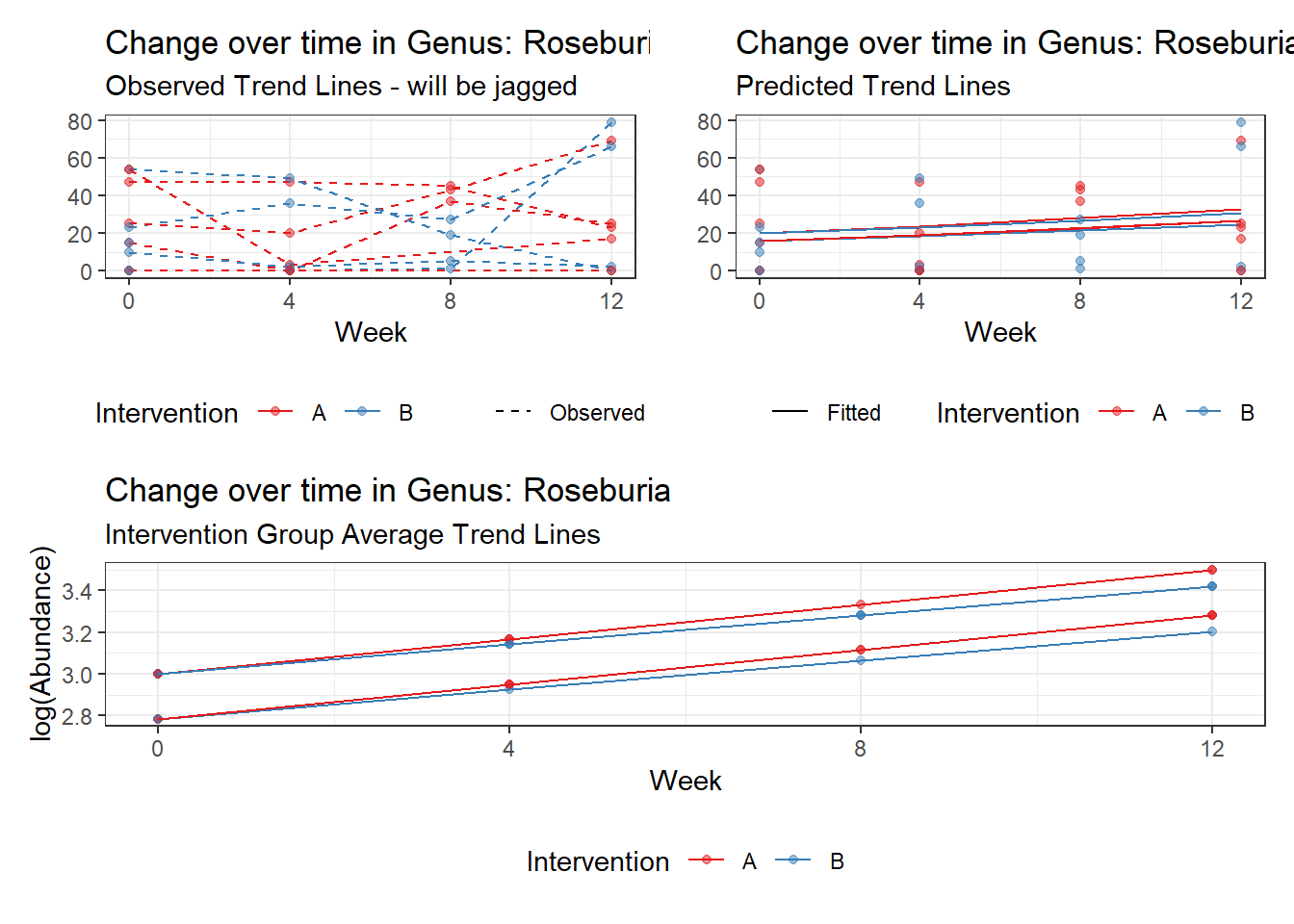
Intraclass Correlation (ICC): 1.191658e-10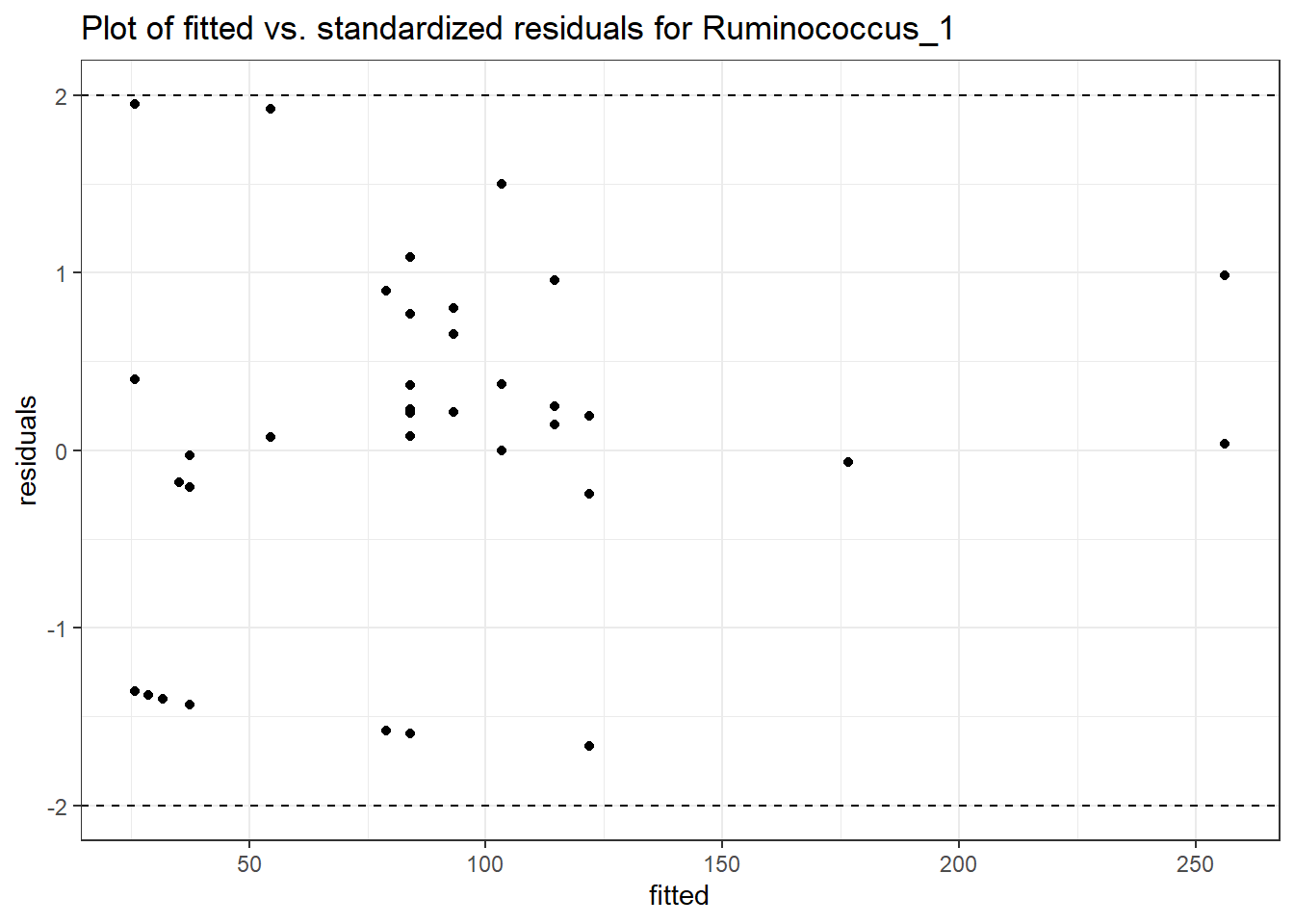
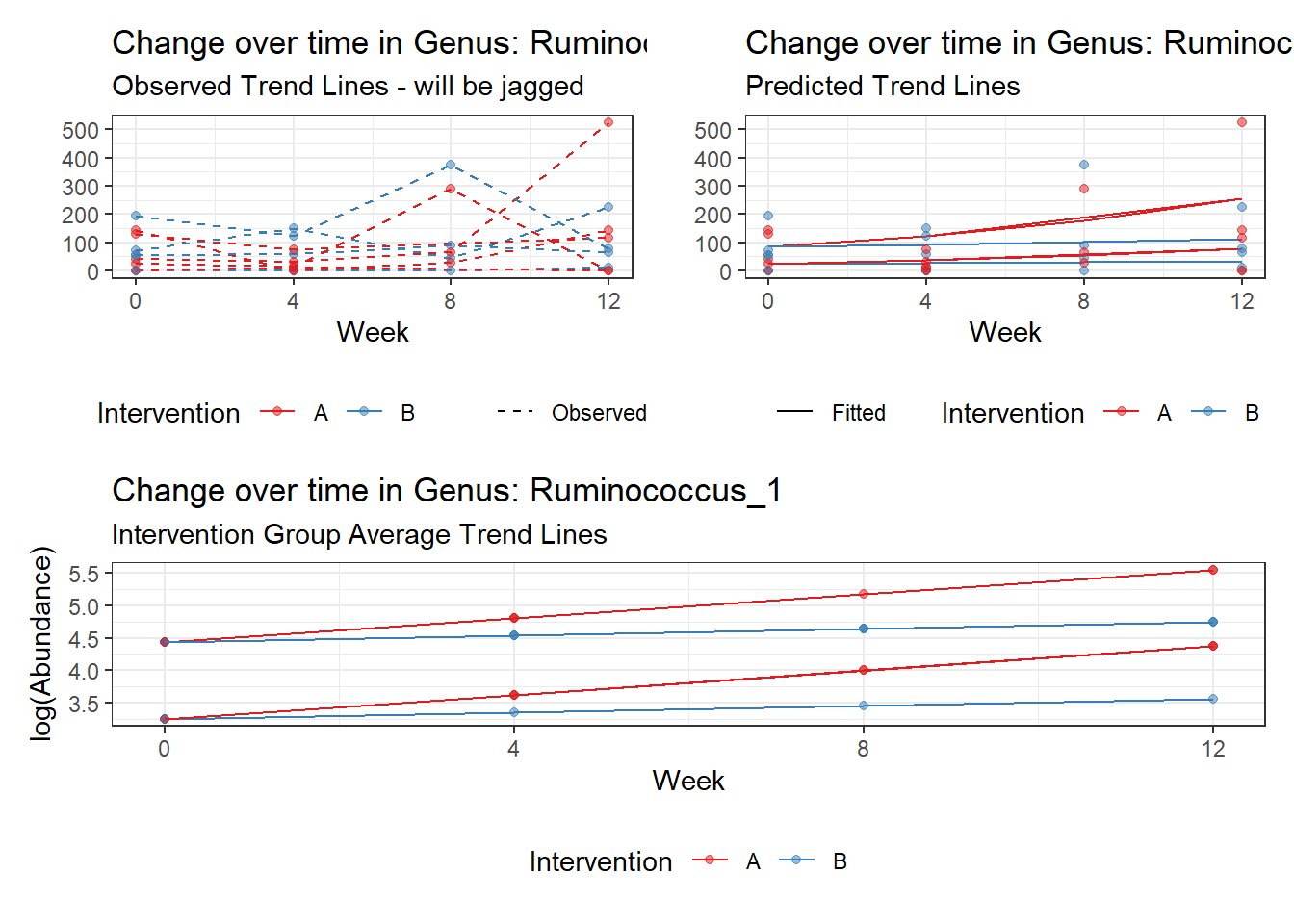
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Ruminococcus_1 3.25397 0.52398 6.21007 0.00000
time Ruminococcus_1 0.37157 0.26396 1.40767 0.15923
hispanic Ruminococcus_1 1.17720 0.61783 1.90538 0.05673
time:intB Ruminococcus_1 -0.26814 0.33574 -0.79865 0.42449
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0916e-05
Significance test not available for random effectsSummary of Effect of Intervention on Abundance
To summarize these results, we are looking 1 main things: that the interaction between intervention and time (intB:time) is significant. If so, then we have evidence that that genus was significantly affected by the intervention, meaning that as time progresses those in the intervention had their microbiome abundance differentially changed over time.
This is sumarized in two ways (i.e., two models). These models are detailed below.
Model 6: Intervention by Time Interaction Only
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\] where, the major aim was to assess the significance of two terms:
- the interaction between time and intervention (intB:time) (\(\gamma_{11}\)) - which assessing the magnitude of the change in alpha diversity as the intervention progresses.
model6.results <- unlist_results(genus.fit6b$`Fixed Effects`)Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")kable(model6.results, format="html", digits=3)%>%
kable_styling(full_width = T) %>%
scroll_box(width="100%",height="5in")| Genus | Term | Estimate | Std. Error | z value | Pr(>|z|) | p.adjust |
|---|---|---|---|---|---|---|
| Akkermansia | time | -0.276 | 0.447 | -0.619 | 0.536 | 0.750 |
| Akkermansia | time:intB | 0.236 | 0.468 | 0.504 | 0.614 | 0.750 |
| Bacteroides | time | -0.031 | 0.058 | -0.545 | 0.586 | 0.750 |
| Bacteroides | time:intB | -0.076 | 0.085 | -0.898 | 0.369 | 0.580 |
| Bifidobacterium | time | -0.304 | 0.315 | -0.965 | 0.334 | 0.580 |
| Bifidobacterium | time:intB | 0.553 | 0.368 | 1.504 | 0.133 | 0.313 |
| Clostridium_sensu_stricto_1 | time | -0.311 | 0.318 | -0.979 | 0.327 | 0.580 |
| Clostridium_sensu_stricto_1 | time:intB | 1.004 | 0.363 | 2.766 | 0.006 | 0.017 |
| Dorea | time | -0.067 | 0.122 | -0.550 | 0.582 | 0.750 |
| Dorea | time:intB | -0.078 | 0.149 | -0.524 | 0.601 | 0.750 |
| Faecalibacterium | time | 0.036 | 0.177 | 0.201 | 0.841 | 0.991 |
| Faecalibacterium | time:intB | -0.170 | 0.183 | -0.932 | 0.351 | 0.580 |
| Lachnospira | time | 0.074 | 0.022 | 3.321 | 0.001 | 0.003 |
| Lachnospira | time:intB | 0.131 | 0.037 | 3.500 | 0.000 | 0.002 |
| Lactobacillus | time | -1.252 | 1.019 | -1.228 | 0.219 | 0.482 |
| Lactobacillus | time:intB | -0.883 | 0.976 | -0.906 | 0.365 | 0.580 |
| Prevotella | time | -0.062 | 0.581 | -0.107 | 0.915 | 0.996 |
| Prevotella | time:intB | 0.003 | 0.613 | 0.005 | 0.996 | 0.996 |
| Roseburia | time | 0.142 | 0.244 | 0.582 | 0.561 | 0.750 |
| Roseburia | time:intB | -0.011 | 0.270 | -0.043 | 0.966 | 0.996 |
| Ruminococcus_1 | time | 0.293 | 0.259 | 1.134 | 0.257 | 0.530 |
| Ruminococcus_1 | time:intB | -0.018 | 0.307 | -0.059 | 0.953 | 0.996 |
Dual results for double check
model6.pois.nb.results <- unlist_dual_results(genus.fit6$`Fixed Effects`, genus.fit6b$`Fixed Effects`)Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Genus", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")kable(model6.pois.nb.results, format="html", digits=3)%>%
kable_styling(full_width = T) %>%
add_header_above(c('Poisson Model'=7, 'Negative Binomial Model'=7)) %>%
scroll_box(width="100%",height="5in")| pois.Genus | pois.Term | pois.Estimate | pois.Std. Error | pois.z value | pois.Pr(>|z|) | pois.p.adjust | nb.Genus | nb.Term | nb.Estimate | nb.Std. Error | nb.z value | nb.Pr(>|z|) | nb.p.adjust |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Akkermansia | time | -0.123 | 0.013 | -9.503 | 0.000 | 0.000 | Akkermansia | time | -0.276 | 0.447 | -0.619 | 0.536 | 0.750 |
| Akkermansia | time:intB | 0.274 | 0.019 | 14.530 | 0.000 | 0.000 | Akkermansia | time:intB | 0.236 | 0.468 | 0.504 | 0.614 | 0.750 |
| Bacteroides | time | -0.023 | 0.004 | -5.451 | 0.000 | 0.000 | Bacteroides | time | -0.031 | 0.058 | -0.545 | 0.586 | 0.750 |
| Bacteroides | time:intB | -0.067 | 0.006 | -10.669 | 0.000 | 0.000 | Bacteroides | time:intB | -0.076 | 0.085 | -0.898 | 0.369 | 0.580 |
| Bifidobacterium | time | -0.121 | 0.062 | -1.969 | 0.049 | 0.065 | Bifidobacterium | time | -0.304 | 0.315 | -0.965 | 0.334 | 0.580 |
| Bifidobacterium | time:intB | 0.263 | 0.076 | 3.471 | 0.001 | 0.001 | Bifidobacterium | time:intB | 0.553 | 0.368 | 1.504 | 0.133 | 0.313 |
| Clostridium_sensu_stricto_1 | time | -0.315 | 0.147 | -2.137 | 0.033 | 0.045 | Clostridium_sensu_stricto_1 | time | -0.311 | 0.318 | -0.979 | 0.327 | 0.580 |
| Clostridium_sensu_stricto_1 | time:intB | 0.688 | 0.162 | 4.241 | 0.000 | 0.000 | Clostridium_sensu_stricto_1 | time:intB | 1.004 | 0.363 | 2.766 | 0.006 | 0.017 |
| Dorea | time | 0.100 | 0.042 | 2.382 | 0.017 | 0.025 | Dorea | time | -0.067 | 0.122 | -0.550 | 0.582 | 0.750 |
| Dorea | time:intB | -0.511 | 0.058 | -8.757 | 0.000 | 0.000 | Dorea | time:intB | -0.078 | 0.149 | -0.524 | 0.601 | 0.750 |
| Faecalibacterium | time | -0.049 | 0.007 | -7.423 | 0.000 | 0.000 | Faecalibacterium | time | 0.036 | 0.177 | 0.201 | 0.841 | 0.991 |
| Faecalibacterium | time:intB | -0.069 | 0.011 | -6.274 | 0.000 | 0.000 | Faecalibacterium | time:intB | -0.170 | 0.183 | -0.932 | 0.351 | 0.580 |
| Lachnospira | time | 0.072 | 0.022 | 3.303 | 0.001 | 0.001 | Lachnospira | time | 0.074 | 0.022 | 3.321 | 0.001 | 0.003 |
| Lachnospira | time:intB | 0.133 | 0.037 | 3.597 | 0.000 | 0.001 | Lachnospira | time:intB | 0.131 | 0.037 | 3.500 | 0.000 | 0.002 |
| Lactobacillus | time | -0.983 | 0.097 | -10.099 | 0.000 | 0.000 | Lactobacillus | time | -1.252 | 1.019 | -1.228 | 0.219 | 0.482 |
| Lactobacillus | time:intB | 0.896 | 0.528 | 1.698 | 0.089 | 0.109 | Lactobacillus | time:intB | -0.883 | 0.976 | -0.906 | 0.365 | 0.580 |
| Prevotella | time | 0.122 | 0.187 | 0.650 | 0.516 | 0.532 | Prevotella | time | -0.062 | 0.581 | -0.107 | 0.915 | 0.996 |
| Prevotella | time:intB | -0.047 | 0.284 | -0.167 | 0.868 | 0.868 | Prevotella | time:intB | 0.003 | 0.613 | 0.005 | 0.996 | 0.996 |
| Roseburia | time | 0.048 | 0.040 | 1.195 | 0.232 | 0.255 | Roseburia | time | 0.142 | 0.244 | 0.582 | 0.561 | 0.750 |
| Roseburia | time:intB | 0.111 | 0.062 | 1.800 | 0.072 | 0.091 | Roseburia | time:intB | -0.011 | 0.270 | -0.043 | 0.966 | 0.996 |
| Ruminococcus_1 | time | 0.411 | 0.023 | 18.062 | 0.000 | 0.000 | Ruminococcus_1 | time | 0.293 | 0.259 | 1.134 | 0.257 | 0.530 |
| Ruminococcus_1 | time:intB | -0.320 | 0.032 | -9.924 | 0.000 | 0.000 | Ruminococcus_1 | time:intB | -0.018 | 0.307 | -0.059 | 0.953 | 0.996 |
write.csv(model6.pois.nb.results, "tab/results_glmm_microbiome_genus.csv")Nice Plot
N <- nrow(microbiome_data$meta.dat)
ids <- microbiome_data$meta.dat$ID
dat <- data.frame(microbiome_data$abund.list[["Genus"]])
# NOTE: Special coding for stripping weird characters from bacteria names
j <- length(rownames(dat))
i <- 1
for(i in 1:j){
while( substring(rownames(dat[i,]), 1, 1) == "_"){
if(i == 1){
row.names(dat) <- c(substring(rownames(dat[i,]), 2),rownames(dat)[2:j])
}
if(i > 1 & i < j){
row.names(dat) <- c(rownames(dat[1:(i-1),]),
substring(rownames(dat[i,]), 2),
rownames(dat)[(i+1):j])
}
if(i == j){
row.names(dat) <- c(rownames(dat[1:(j-1),]),substring(rownames(dat[j,]), 2))
}
} # End while loop
} # End for loop
# ====================== #
num.bact <- nrow(dat)
dat <- t(dat[1:num.bact,1:N])
dat <- apply(dat, 2, function(x){log(x+1)})
k <- ncol(microbiome_data$meta.dat) # number of original variables
dat <- data.frame(cbind(microbiome_data$meta.dat, dat[ids,]))
# first genus
PH <- names(genus.fit6b$`Fitted Models`)
pL <- list()
i <- 1
for(i in 1:length(PH)){
fit <- genus.fit6b$`Fitted Models`[[PH[i]]]
idat <- cbind(dat, fit=exp(predict(fit)))
idat <- idat %>% mutate(Week = (as.numeric(Week)-1)*4)
idat$PRED <- predict(fit, re.form=NA)
idat$Outcome <- idat[, PH[i]]
# extract p-value
pv <- filter(model6.results, Genus == PH[i], Term == "time:intB")
pv <- ifelse(pv[7] >= .001, sprintf("= %.3f", pv[7]), "< 0.001")
# fix genus names:
if(PH[i] == "Clostridium_sensu_stricto_1") PH[i] <- "Clostridium"
if(PH[i] == "Ruminococcus_1") PH[i] <- "Ruminococcus"
pL[[PH[i]]] <- ggplot(idat, aes(Week, PRED, color=Intervention,
group=SubjectID))+
geom_line() +
#geom_jitter(aes(x=Week, y=Outcome),
# width=0.25, height = 0)+
scale_x_continuous(breaks=c(0,4,8,12))+
scale_y_continuous(limits=c(-5,10), breaks=seq(-5,10,2.5))+
labs(y="log(Abundance)", title=paste0(PH[i])) +
annotate("text", x=6, y=10, label=paste0("p ", pv))+
theme_classic()+
theme(legend.position = c(0.5,0.5))
}
# extract legend
leg <- get_legend(pL[[1]])
# some specializing for more space
pL[[1]] <- pL[[1]] + theme(legend.position = "none",
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
pL[[2]] <- pL[[2]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[3]] <- pL[[3]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[4]] <- pL[[4]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[5]] <- pL[[5]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[6]] <- pL[[6]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[7]] <- pL[[7]] + theme(legend.position = "none")
pL[[8]] <- pL[[8]] + theme(legend.position = "none",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())
pL[[9]] <- pL[[9]] + theme(legend.position = "none",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())
pL[[10]] <- pL[[10]] + theme(legend.position = "none",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())
pL[[11]] <- pL[[11]] + theme(legend.position = "none",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())
# merge plot
p <- (pL[[1]] + pL[[2]] + pL[[3]] + pL[[4]] + pL[[5]] + pL[[6]] + pL[[7]] + pL[[8]] + pL[[9]] + pL[[10]] + pL[[11]] + leg) + plot_layout(ncol=6, nrow=2)
p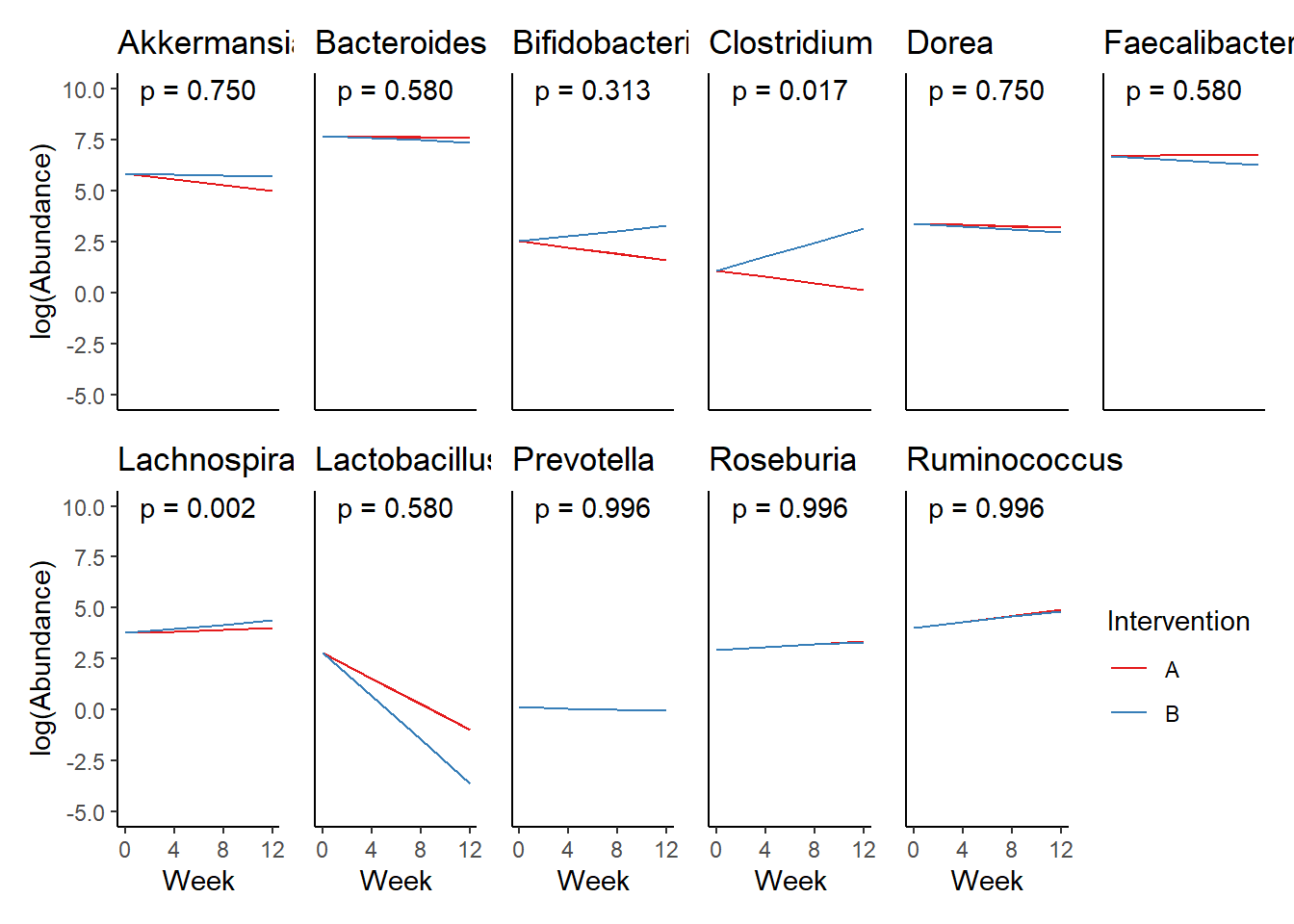
#ggsave("fig/figure5_glmm_genus.pdf", p, units="in", width=10, height=4)
sessionInfo()R version 3.6.3 (2020-02-29)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 18362)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.0.0 microbiome_1.8.0 car_3.0-8 carData_3.0-4
[5] gvlma_1.0.0.3 patchwork_1.0.0 viridis_0.5.1 viridisLite_0.3.0
[9] gridExtra_2.3 xtable_1.8-4 kableExtra_1.1.0 plyr_1.8.6
[13] data.table_1.12.8 readxl_1.3.1 forcats_0.5.0 stringr_1.4.0
[17] dplyr_0.8.5 purrr_0.3.4 readr_1.3.1 tidyr_1.1.0
[21] tibble_3.0.1 ggplot2_3.3.0 tidyverse_1.3.0 lmerTest_3.1-2
[25] lme4_1.1-23 Matrix_1.2-18 vegan_2.5-6 lattice_0.20-38
[29] permute_0.9-5 phyloseq_1.30.0
loaded via a namespace (and not attached):
[1] Rtsne_0.15 minqa_1.2.4 colorspace_1.4-1
[4] rio_0.5.16 ellipsis_0.3.1 rprojroot_1.3-2
[7] XVector_0.26.0 fs_1.4.1 rstudioapi_0.11
[10] farver_2.0.3 fansi_0.4.1 lubridate_1.7.8
[13] xml2_1.3.2 codetools_0.2-16 splines_3.6.3
[16] knitr_1.28 ade4_1.7-15 jsonlite_1.6.1
[19] workflowr_1.6.2 nloptr_1.2.2.1 broom_0.5.6
[22] cluster_2.1.0 dbplyr_1.4.4 BiocManager_1.30.10
[25] compiler_3.6.3 httr_1.4.1 backports_1.1.7
[28] assertthat_0.2.1 cli_2.0.2 later_1.0.0
[31] htmltools_0.4.0 tools_3.6.3 igraph_1.2.5
[34] gtable_0.3.0 glue_1.4.1 reshape2_1.4.4
[37] Rcpp_1.0.4.6 Biobase_2.46.0 cellranger_1.1.0
[40] vctrs_0.3.0 Biostrings_2.54.0 multtest_2.42.0
[43] ape_5.3 nlme_3.1-144 iterators_1.0.12
[46] xfun_0.14 openxlsx_4.1.5 rvest_0.3.5
[49] lifecycle_0.2.0 statmod_1.4.34 zlibbioc_1.32.0
[52] MASS_7.3-51.5 scales_1.1.1 hms_0.5.3
[55] promises_1.1.0 parallel_3.6.3 biomformat_1.14.0
[58] rhdf5_2.30.1 RColorBrewer_1.1-2 curl_4.3
[61] yaml_2.2.1 stringi_1.4.6 highr_0.8
[64] S4Vectors_0.24.4 foreach_1.5.0 BiocGenerics_0.32.0
[67] zip_2.0.4 boot_1.3-24 rlang_0.4.6
[70] pkgconfig_2.0.3 evaluate_0.14 Rhdf5lib_1.8.0
[73] labeling_0.3 tidyselect_1.1.0 magrittr_1.5
[76] R6_2.4.1 IRanges_2.20.2 generics_0.0.2
[79] DBI_1.1.0 foreign_0.8-75 pillar_1.4.4
[82] haven_2.3.0 withr_2.2.0 mgcv_1.8-31
[85] abind_1.4-5 survival_3.1-8 modelr_0.1.8
[88] crayon_1.3.4 rmarkdown_2.1 grid_3.6.3
[91] blob_1.2.1 git2r_0.27.1 reprex_0.3.0
[94] digest_0.6.25 webshot_0.5.2 httpuv_1.5.2
[97] numDeriv_2016.8-1.1 stats4_3.6.3 munsell_0.5.0