Modeling Changes in Microbiome
Last updated: 2020-06-17
Checks: 6 1
Knit directory: Fiber_Intervention_Study/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a16e6ef. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/.Rhistory
Ignored: reference-papers/Dietary_Variables.xlsx
Ignored: reference-papers/Johnson_2019.pdf
Ignored: renv/library/
Ignored: renv/staging/
Untracked files:
Untracked: analysis/glme_microbiome_genus_subset.Rmd
Untracked: analysis/glme_microbiome_phylum.Rmd
Untracked: data/analysis-data/DataDictionary_TOTALS_2018Record.xls
Untracked: fig/figure5_glmm_genus.pdf
Untracked: fig/figure5_glmm_phylum.pdf
Untracked: fig/figure5_glmm_phylum.png
Untracked: tab/results_glmm_microbiome_genus.csv
Untracked: tab/results_glmm_microbiome_phylum.csv
Untracked: tab/tables_results_2020-06-08.zip
Untracked: tab/tables_results_2020-06-08/
Unstaged changes:
Modified: analysis/data_processing.Rmd
Deleted: analysis/glme_microbiome.Rmd
Modified: analysis/index.Rmd
Modified: analysis/microbiome_diet_trends.Rmd
Modified: code/get_cleaned_data.R
Modified: code/microbiome_statistics_and_functions.R
Modified: fig/figure2.pdf
Modified: fig/figure3.pdf
Modified: fig/figure4.pdf
Modified: fig/figure4_final_microbiome_diet_variables_over_time.pdf
Modified: fig/figure4_legend.pdf
Modified: fig/figure5_glmm.pdf
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
This page contains the investigation of the changes over time.
Data prep
This is only to recode some variables to ease interpretation.
microbiome_data$meta.dat <- microbiome_data$meta.dat %>%
mutate(intB = ifelse(Intervention=="B", 1,0),
time = as.numeric(Week) - 1,
female = ifelse(Gender == "F", 1, 0),
hispanic = ifelse(Ethnicity %in% c("White", "Asian", "Native America"), 1, 0))Phylum Level Microbiome and Intervention
We will build up to the effect of the intervention by investigating covariates to see if we can exclude them from model with intervention. Due to only having 11 people, we will only estimate 1 random effect per model (random intercepts). All other effects are fixed, but we will only estimate up to 2 effects. Meaning we can only include two covariates in each model at a time.
In all models, \(h(.)\) is an arbitrary link function. We selected the Poisson model for the outcomes so the link function is the log link.
For all significance test, we will use \(p < 0.05\) as the level to declare significance, but we will largely focus on magnitude of effects.
Unconditional Model
This is a random intercepts model (worst case). No covariates are included.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \end{align*}\]
phylum.fit0 <- glmm_microbiome(mydata=microbiome_data,model.number=0,
taxa.level="Phylum", link="poisson",
model="1 + (1|SubjectID)")
#########################################
#########################################
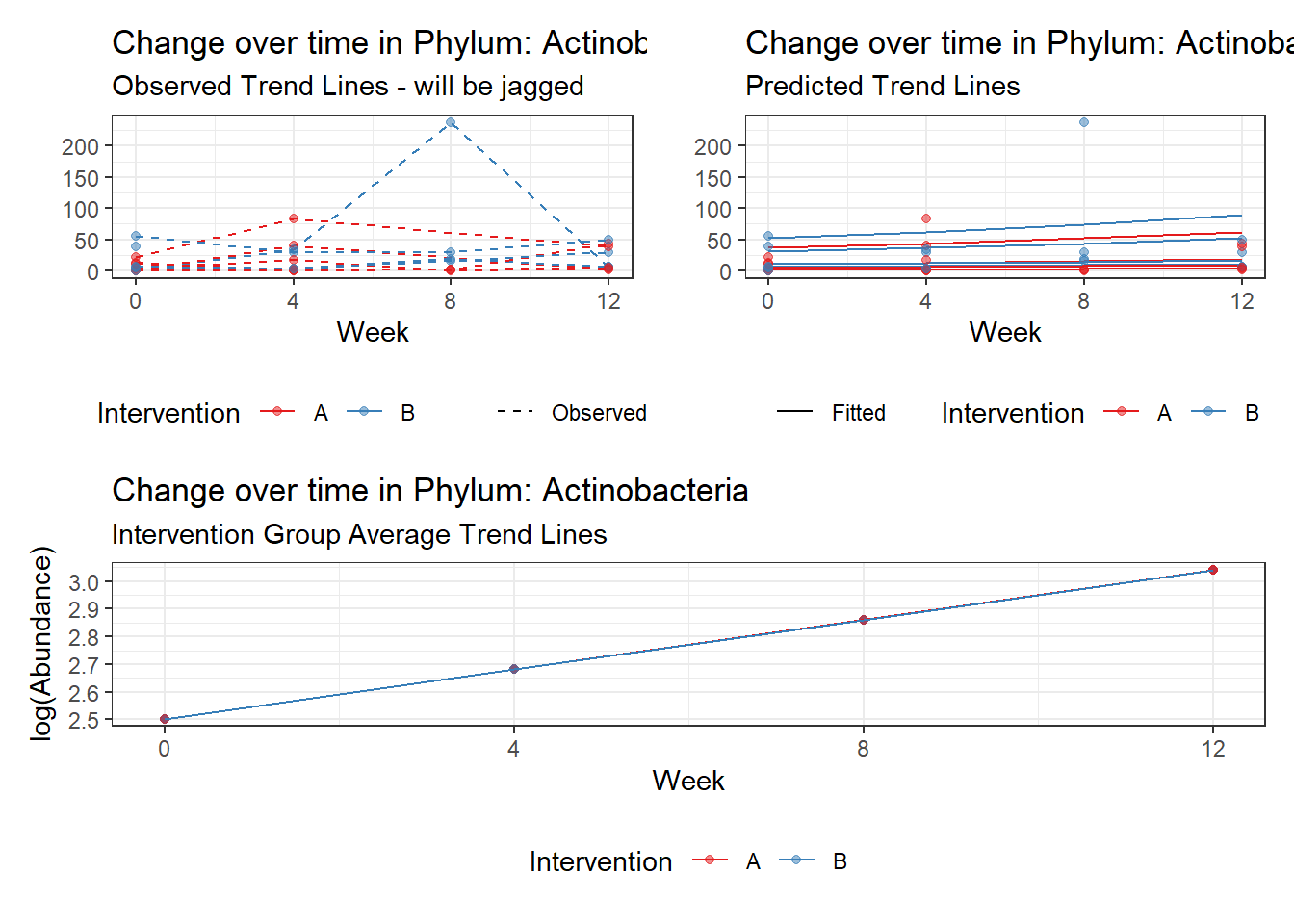
Model: Actinobacteria ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poisson
Intraclass Correlation (ICC): 0.4905836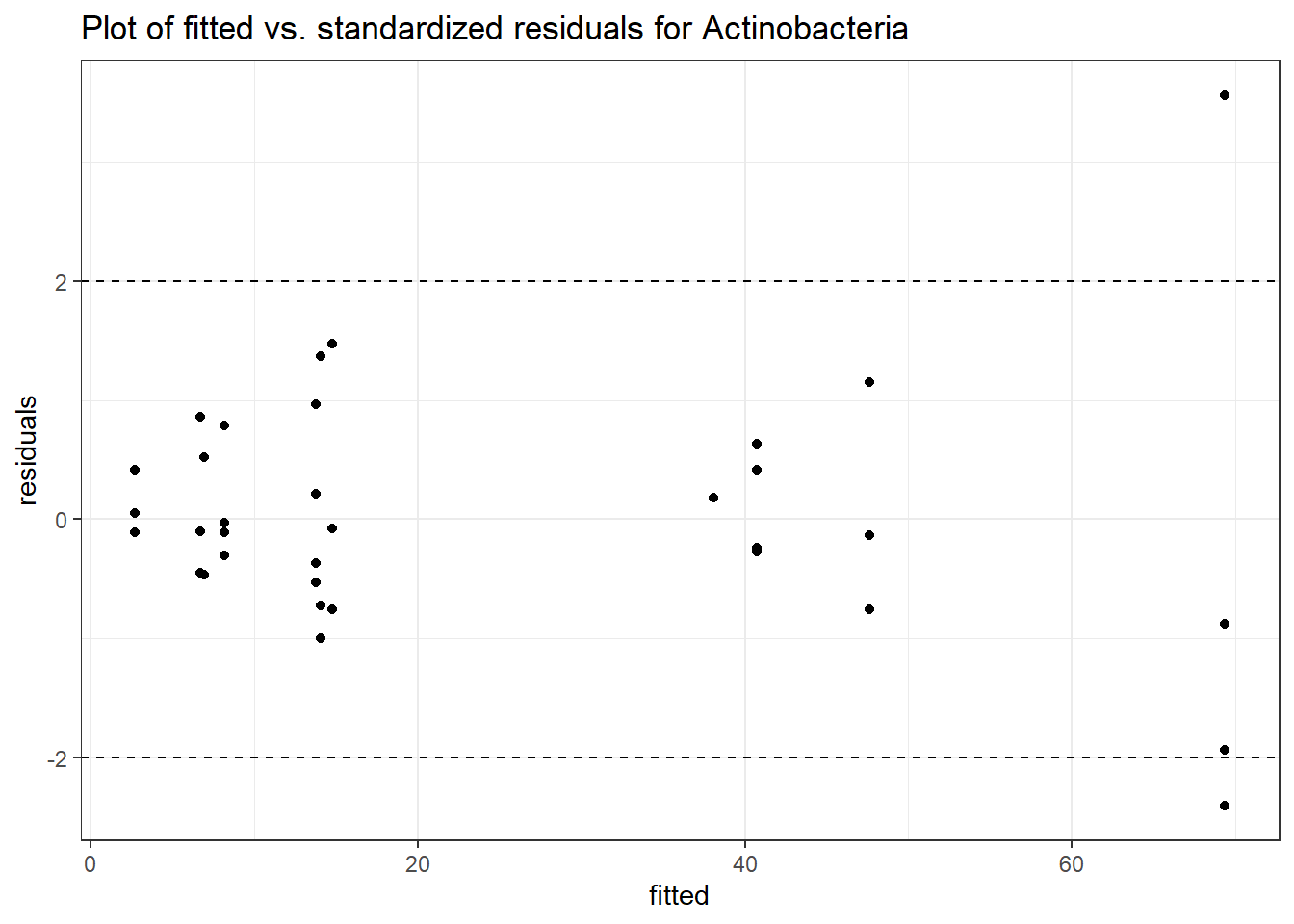
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.74459 0.30057 9.13131 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.98134
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poisson
Intraclass Correlation (ICC): 0.1771389
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 7.97 0.13995 56.94866 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.46397
Significance test not available for random effects
#########################################
#########################################
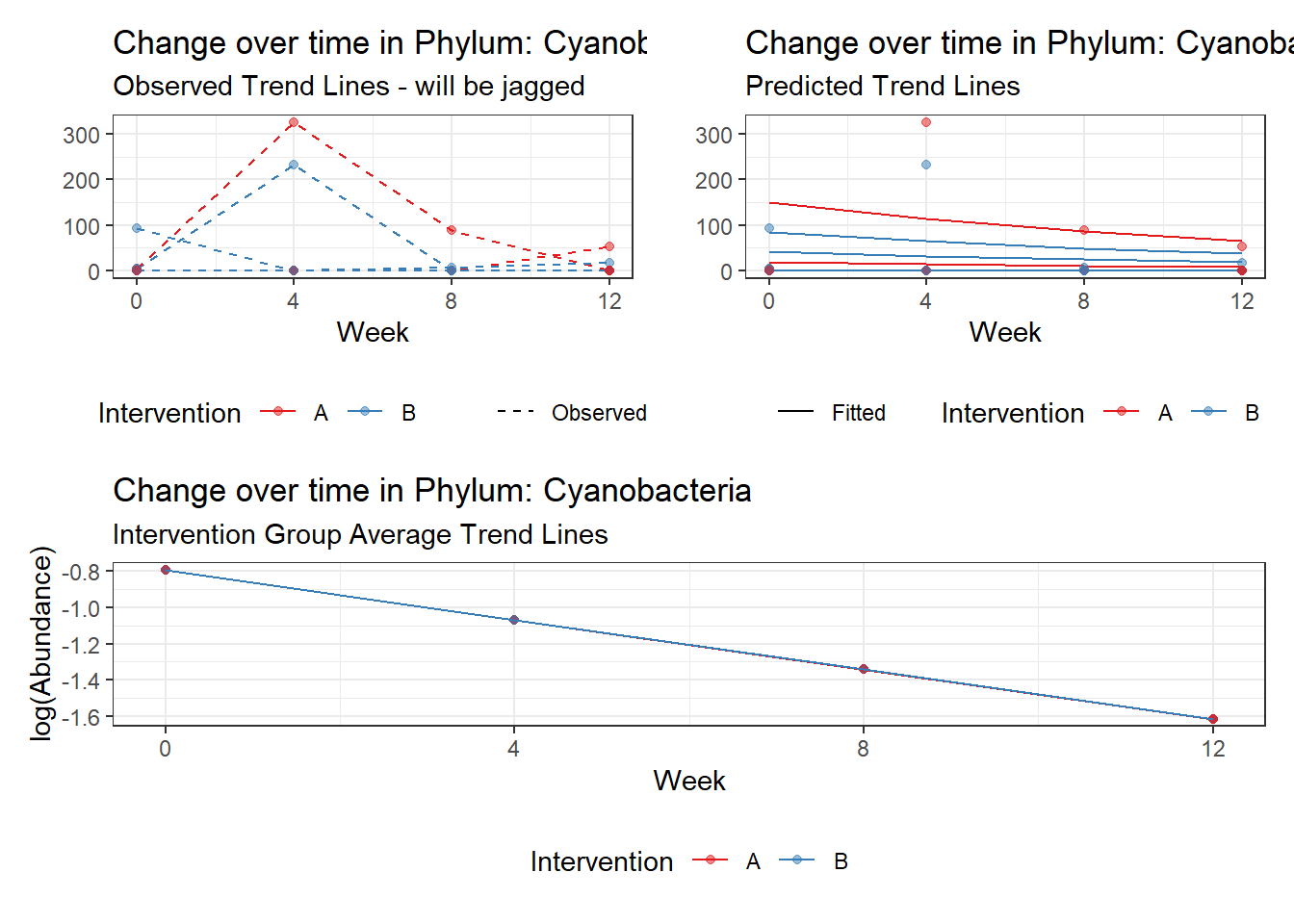
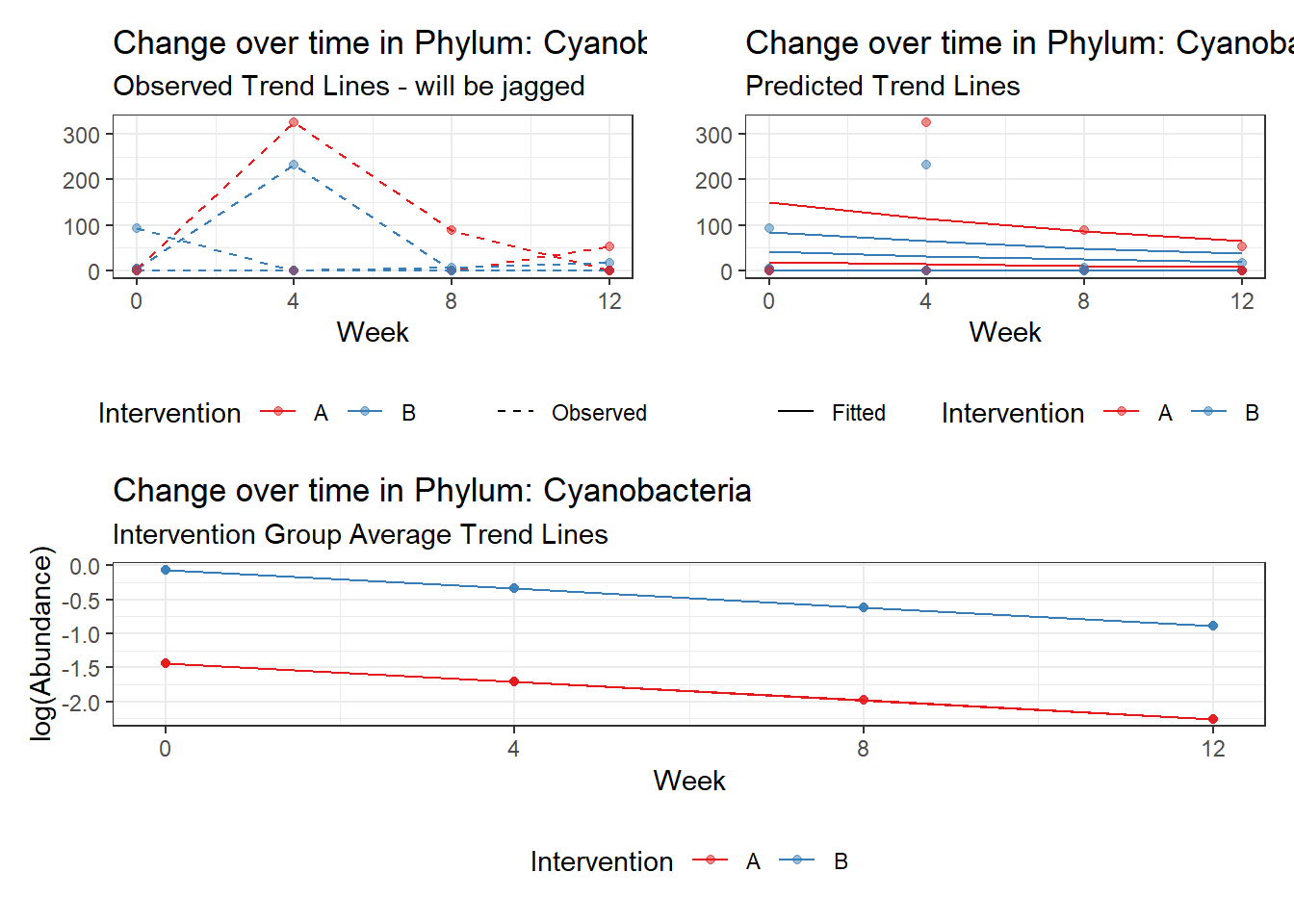
Model: Cyanobacteria ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poisson
Intraclass Correlation (ICC): 0.9437219

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -1.06818 1.6666 -0.64093 0.52156
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.095
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poisson
Intraclass Correlation (ICC): 0.8160485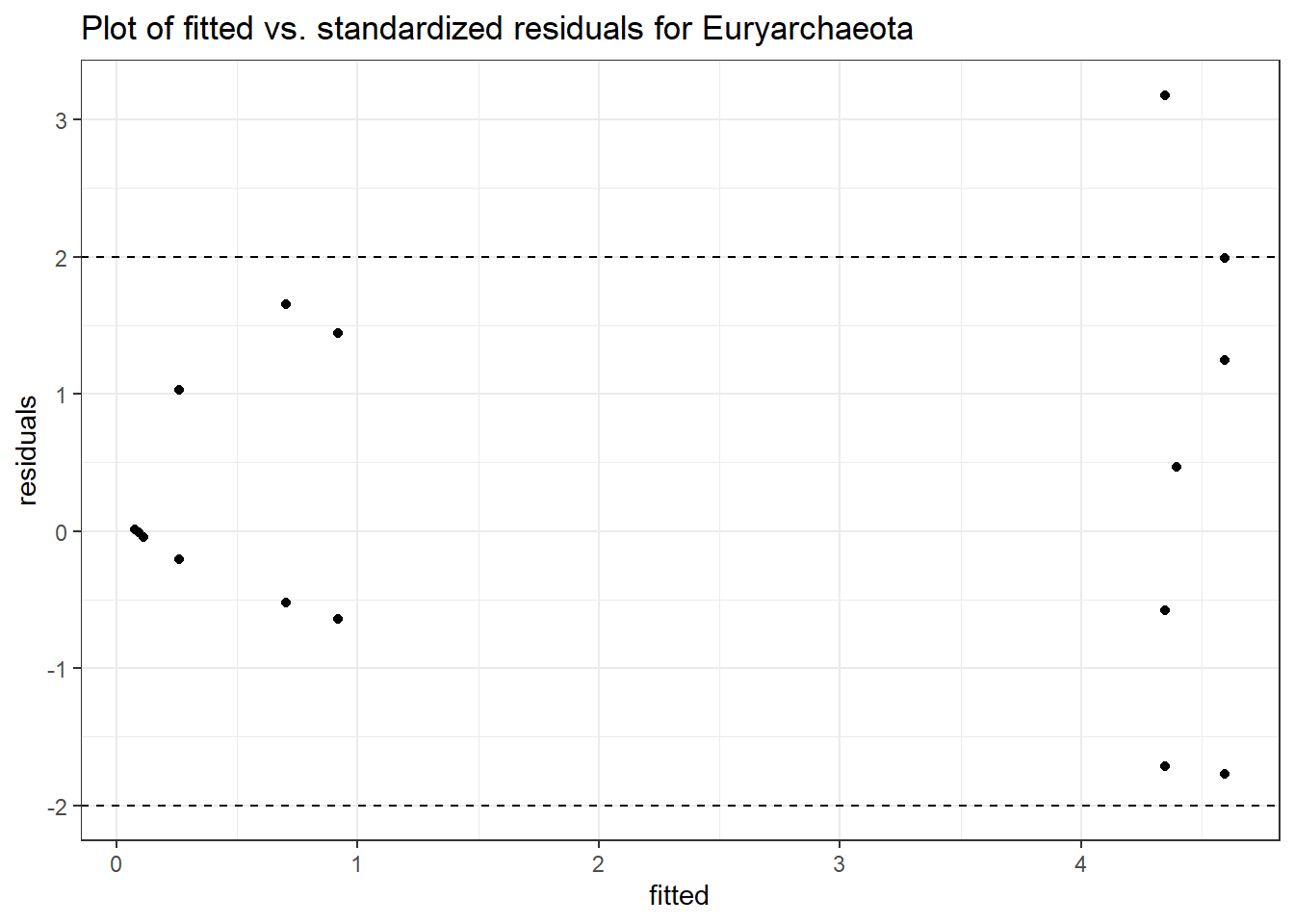

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -1.19078 0.86784 -1.37213 0.17002
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1062
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poisson
Intraclass Correlation (ICC): 0.1341399
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.95503 0.11873 67.00139 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.3936
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poissonWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0470305 (tol = 0.002, component 1)
Intraclass Correlation (ICC): 0.8560092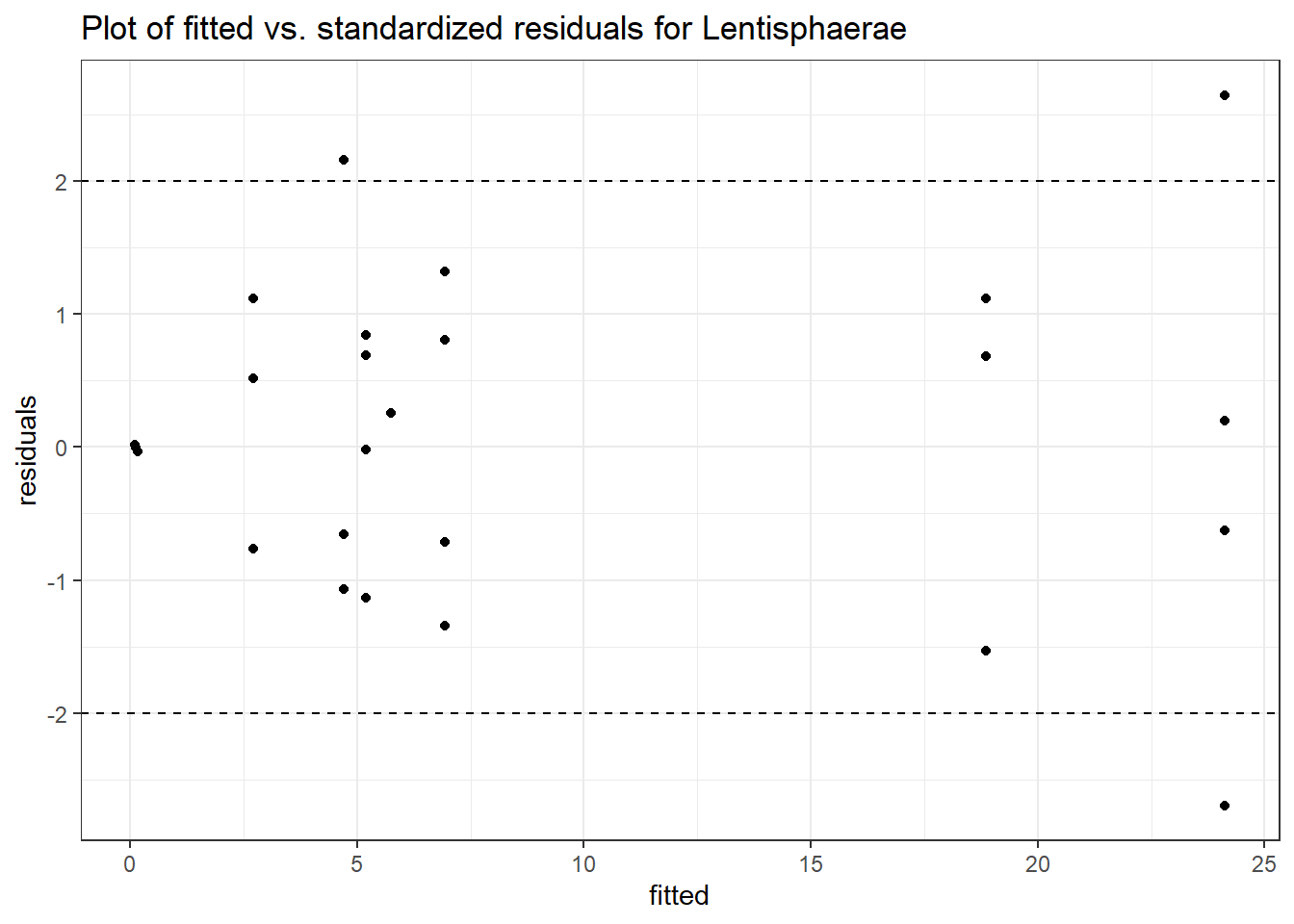

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.27284 0.0015 181.4693 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4382
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poisson
Intraclass Correlation (ICC): 0.08266202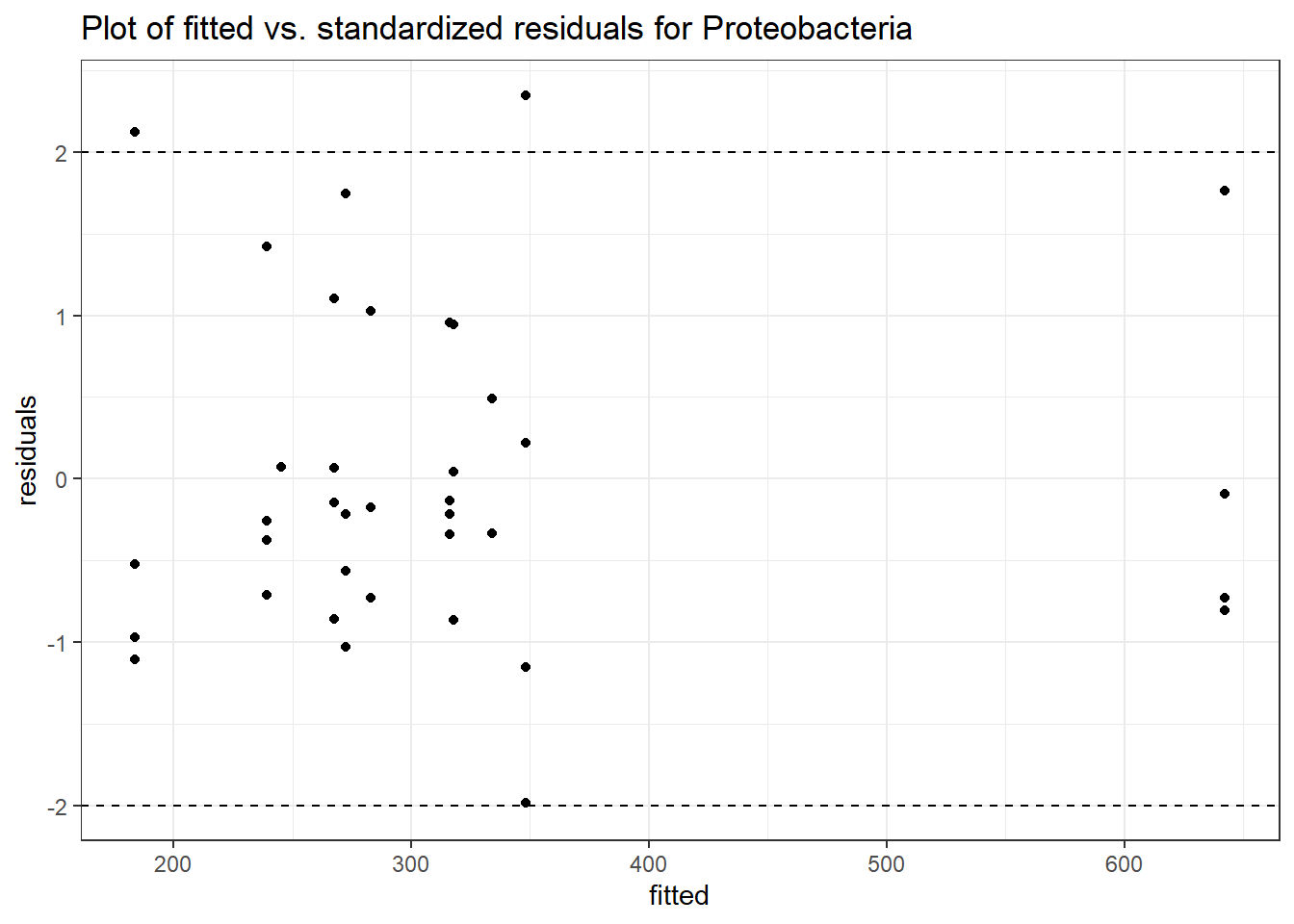

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.69779 0.09114 62.51624 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.30018
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poisson
Intraclass Correlation (ICC): 0.9143442
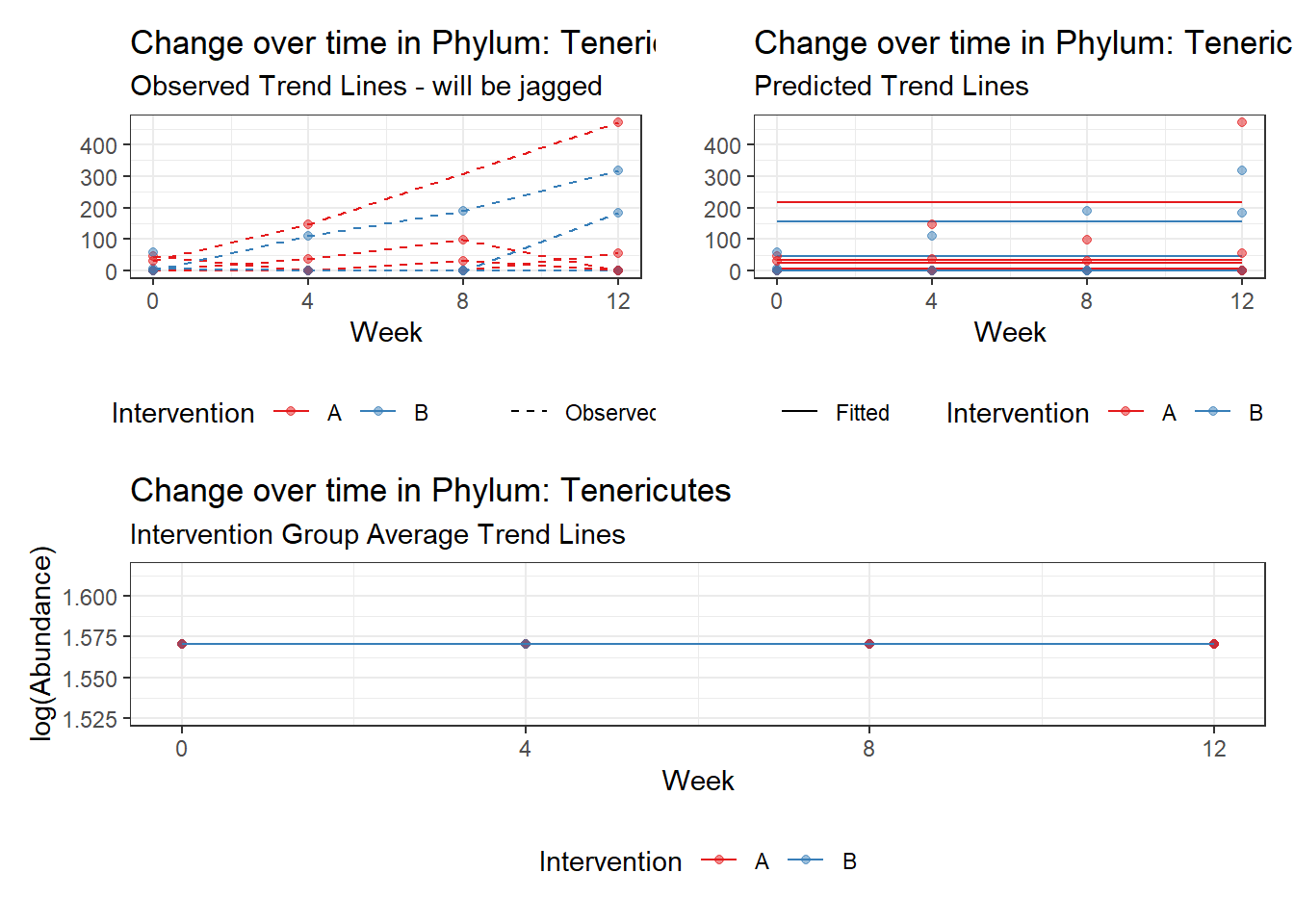
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 1.57029 1.02318 1.53472 0.12485
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.2672
Significance test not available for random effects
#########################################
#########################################
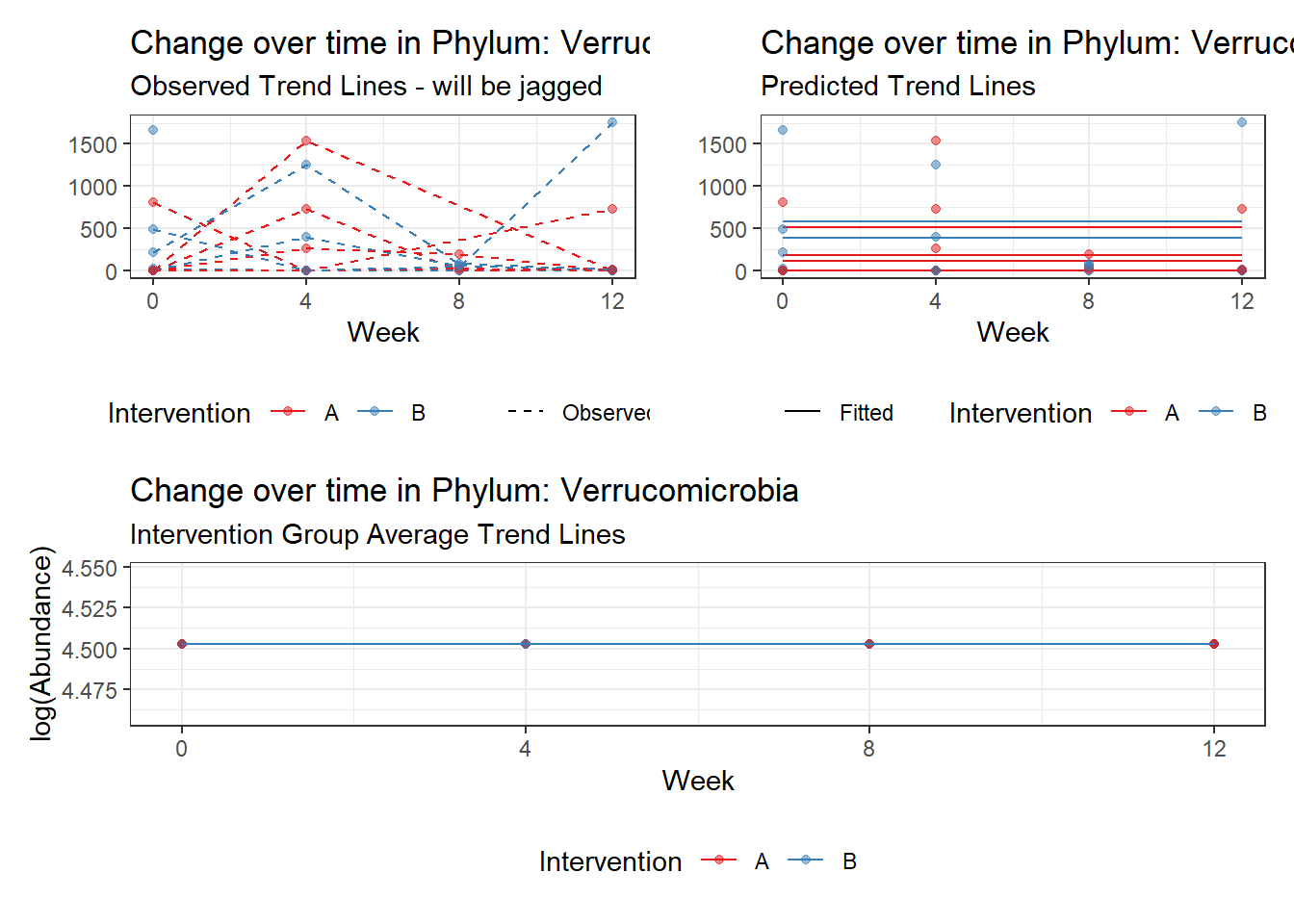
Model: Verrucomicrobia ~ 1 + (1 | SubjectID)
<environment: 0x000000003fc897b0>
Link: poisson
Intraclass Correlation (ICC): 0.8607519
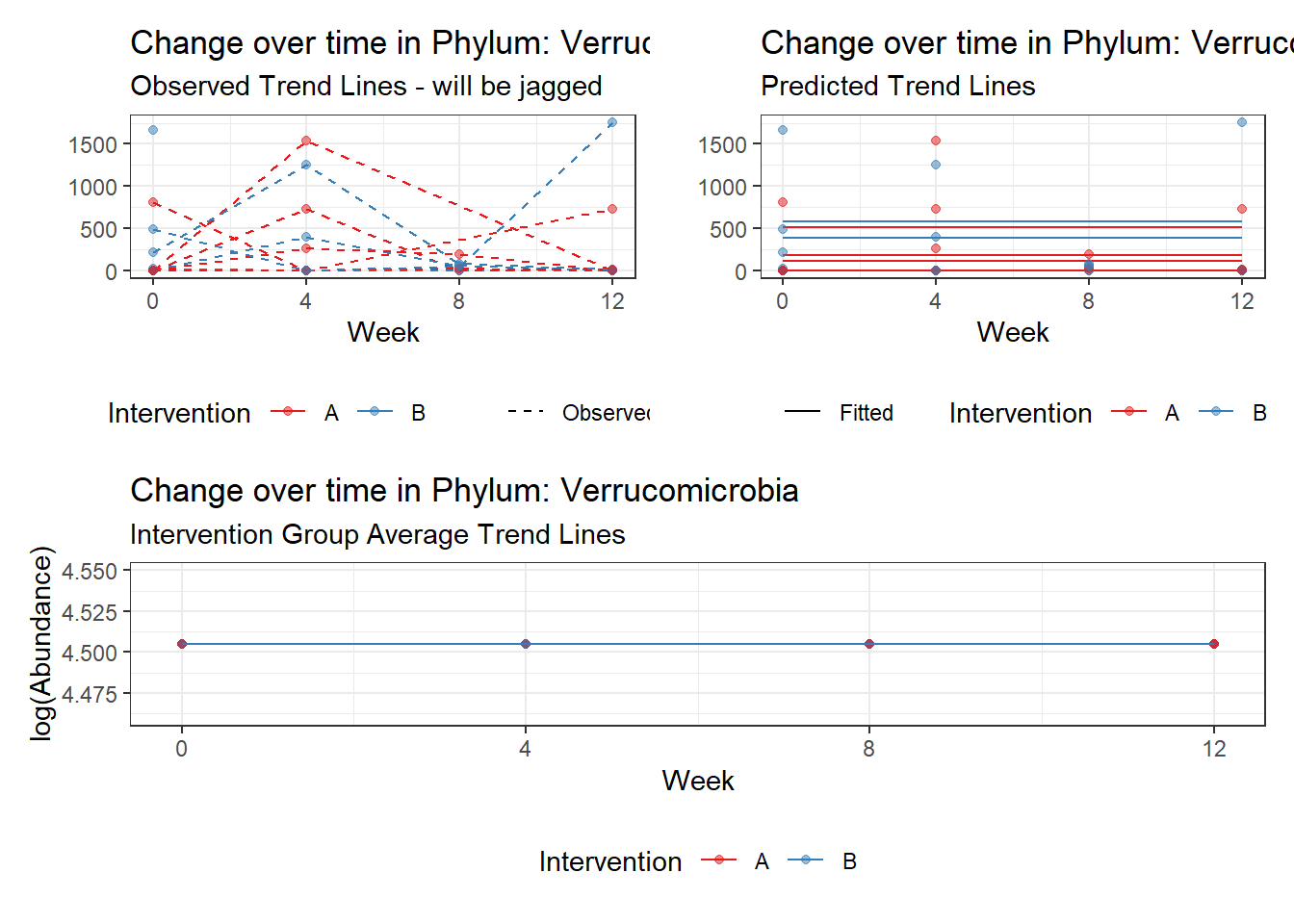
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.50476 0.75627 5.95654 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4862
Significance test not available for random effectsphylum.fit0b <- glmm_microbiome(mydata=microbiome_data,model.number=0,
taxa.level="Phylum", link="negbinom",
model="1 + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinom
Intraclass Correlation (ICC): 0.3549161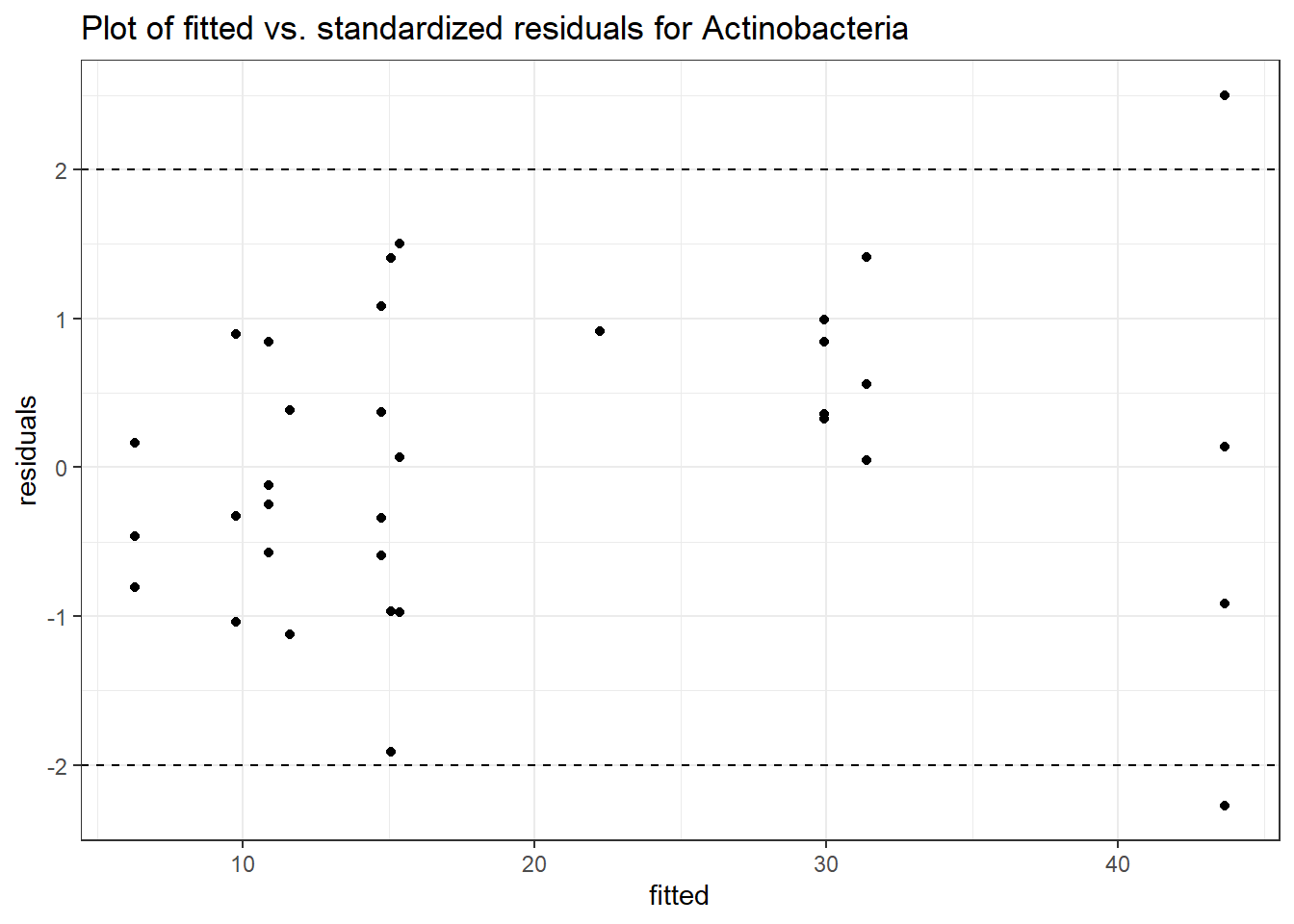

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.79811 0.31298 8.94033 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.74175
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinom
Intraclass Correlation (ICC): 0.1323578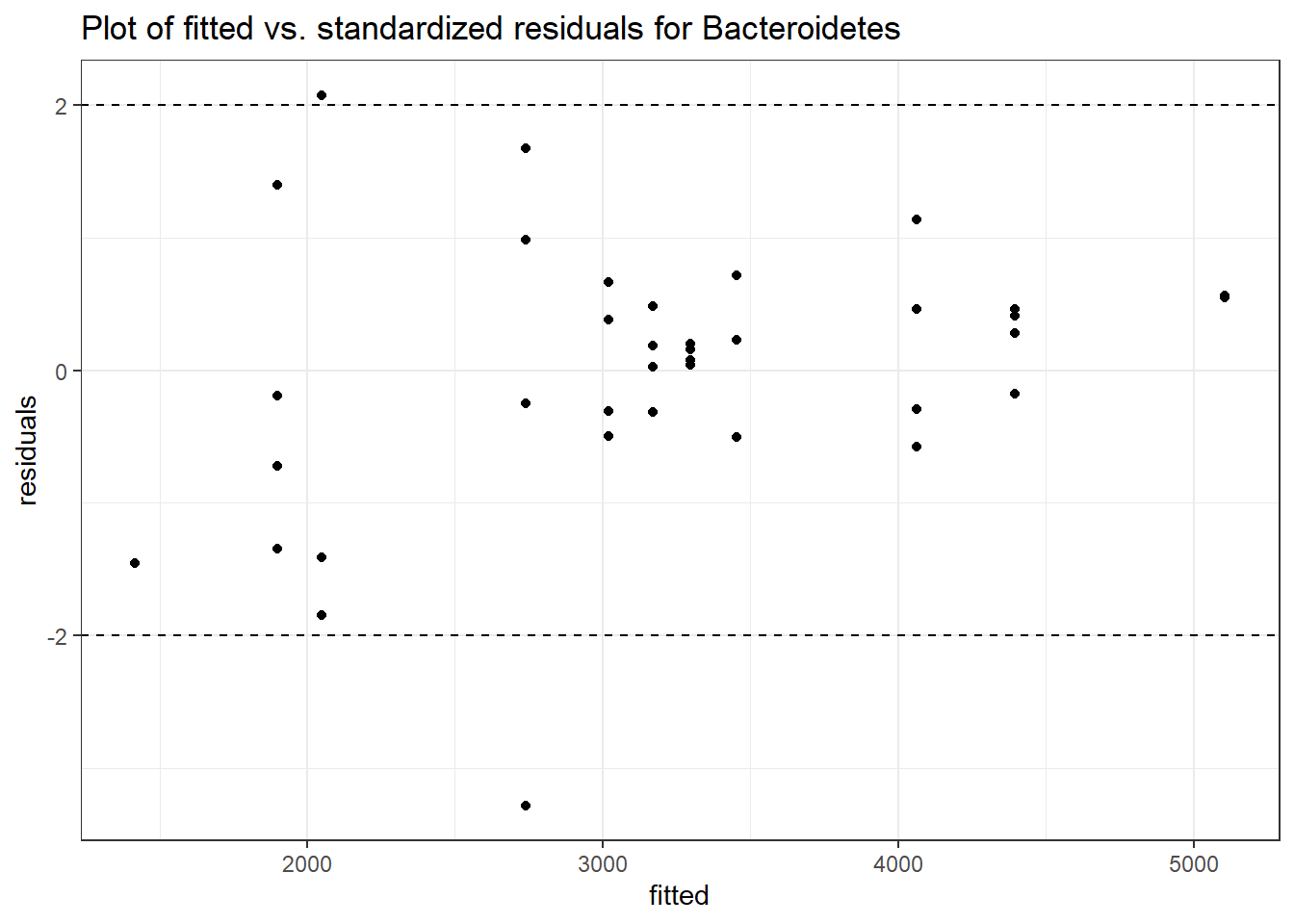
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 7.99127 0.12634 63.25006 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.39057
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced
Intraclass Correlation (ICC): 0.9438998
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -1.07647 1.65978 -0.64856 0.51662
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.1019
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 0
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota 0.2809 0.56099 0.50072 0.61657
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinom
Intraclass Correlation (ICC): 0.12232
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.95573 0.1162 68.46622 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.37332
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0470305 (tol = 0.002, component 1)
Intraclass Correlation (ICC): 0.8198321

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.39317 0.85549 0.45959 0.64581
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1332
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinom
Intraclass Correlation (ICC): 0.02528622
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.7389 0.11704 49.03538 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.16107
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reached
Warning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, : NaNs
produced
Intraclass Correlation (ICC): 0.915808
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 1.54525 1.07089 1.44296 0.14903
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.2981
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + (1 | SubjectID)
<environment: 0x000000004e7b2968>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reached
Warning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, : NaNs
produced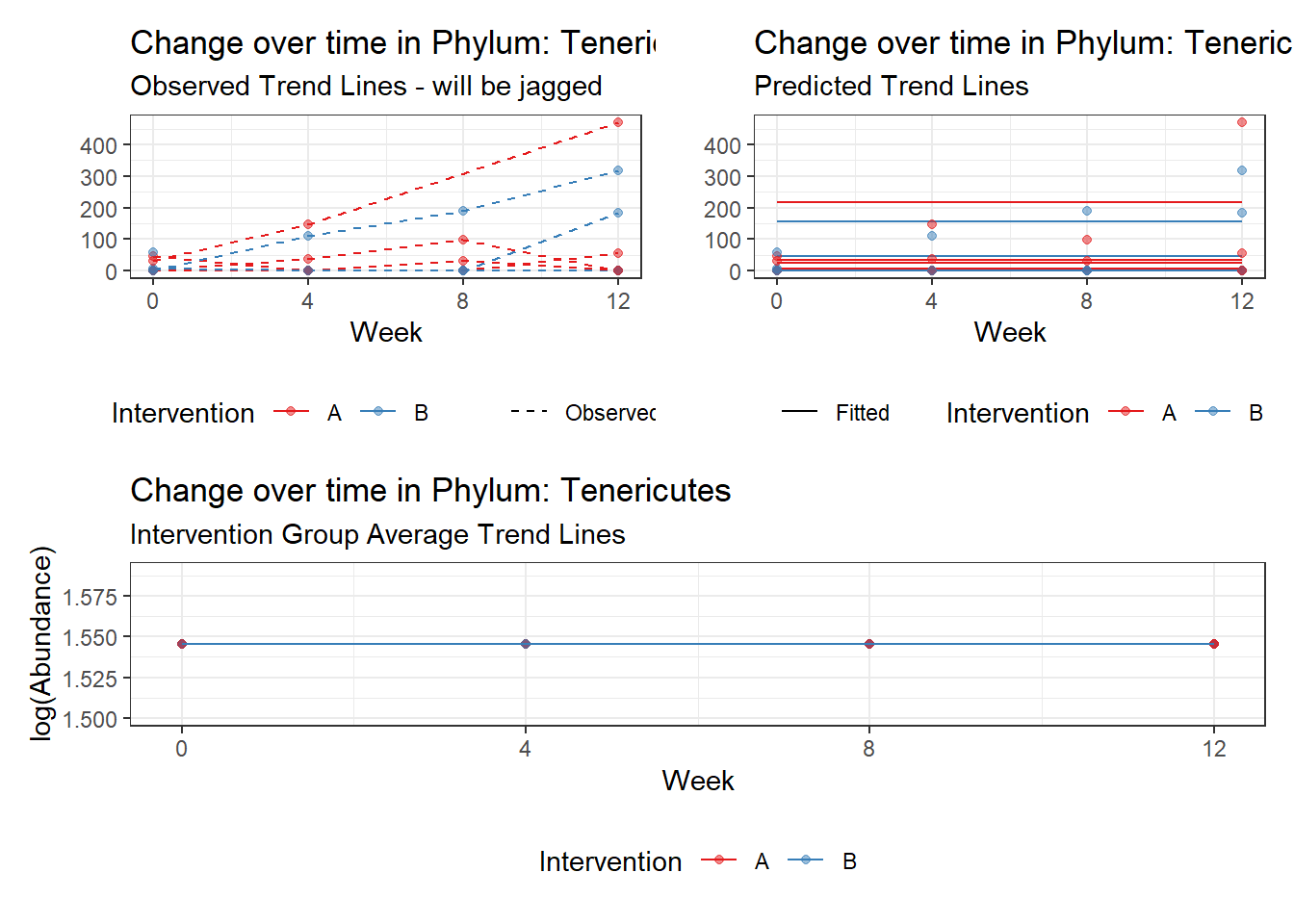
Intraclass Correlation (ICC): 0.8611513
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.50251 0.75647 5.95201 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4904
Significance test not available for random effectsFixed Effect of Time
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
phylum.fit1 <- glmm_microbiome(mydata=microbiome_data,model.number=1,
taxa.level="Phylum", link="poisson",
model="1 + time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.4983818
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.50315 0.30833 8.11831 0
time Actinobacteria 0.17905 0.03052 5.86651 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.99677
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.1810136

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.00956 0.14184 56.46826 0
time Bacteroidetes -0.03231 0.00265 -12.17927 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.47013
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.944816
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -0.79541 1.68718 -0.47144 0.63732
time Cyanobacteria -0.27255 0.03201 -8.51538 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.1378
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.8154851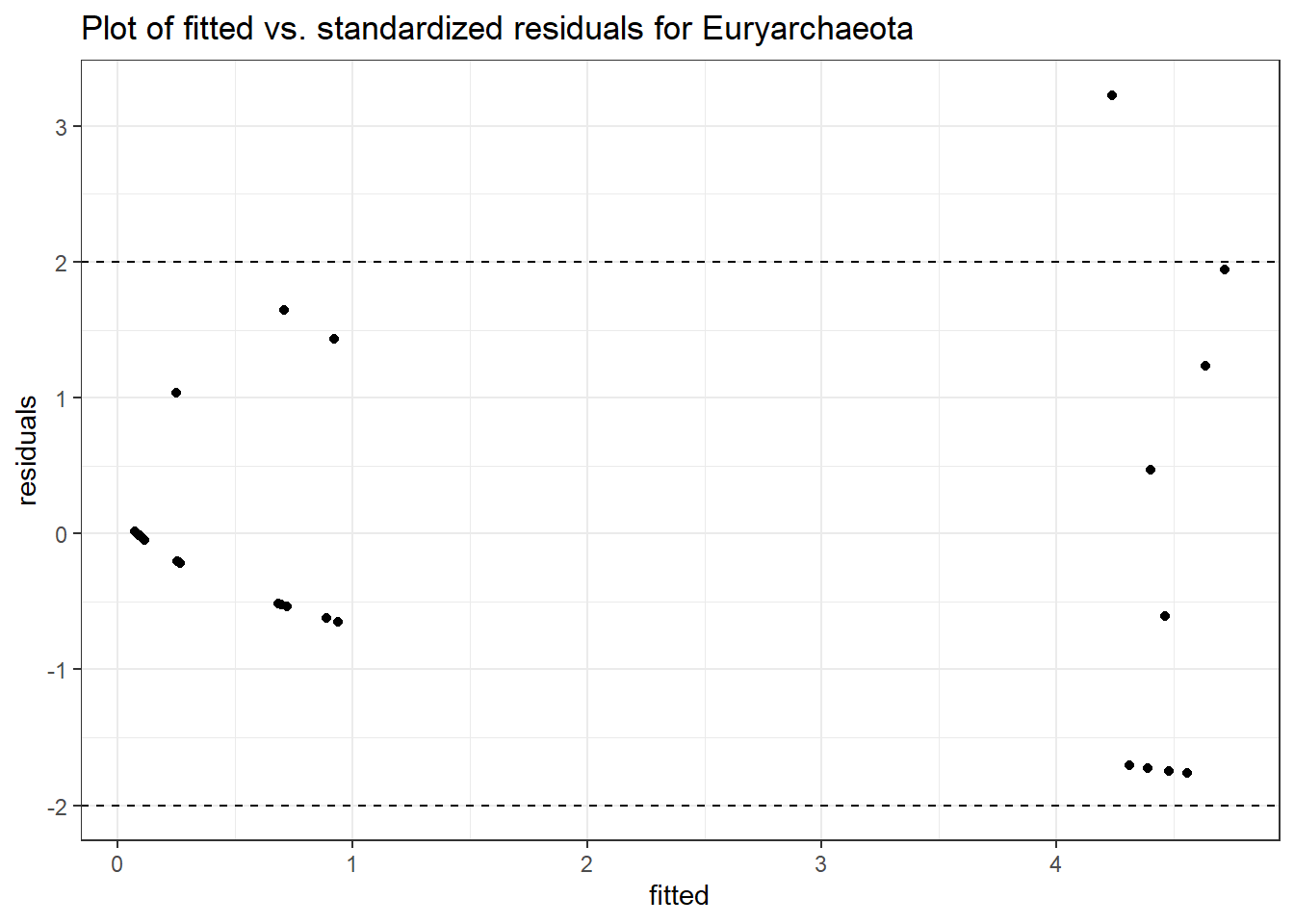
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -1.16699 0.88479 -1.31895 0.18718
time Euryarchaeota -0.01744 0.13284 -0.13128 0.89555
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1023
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.1323942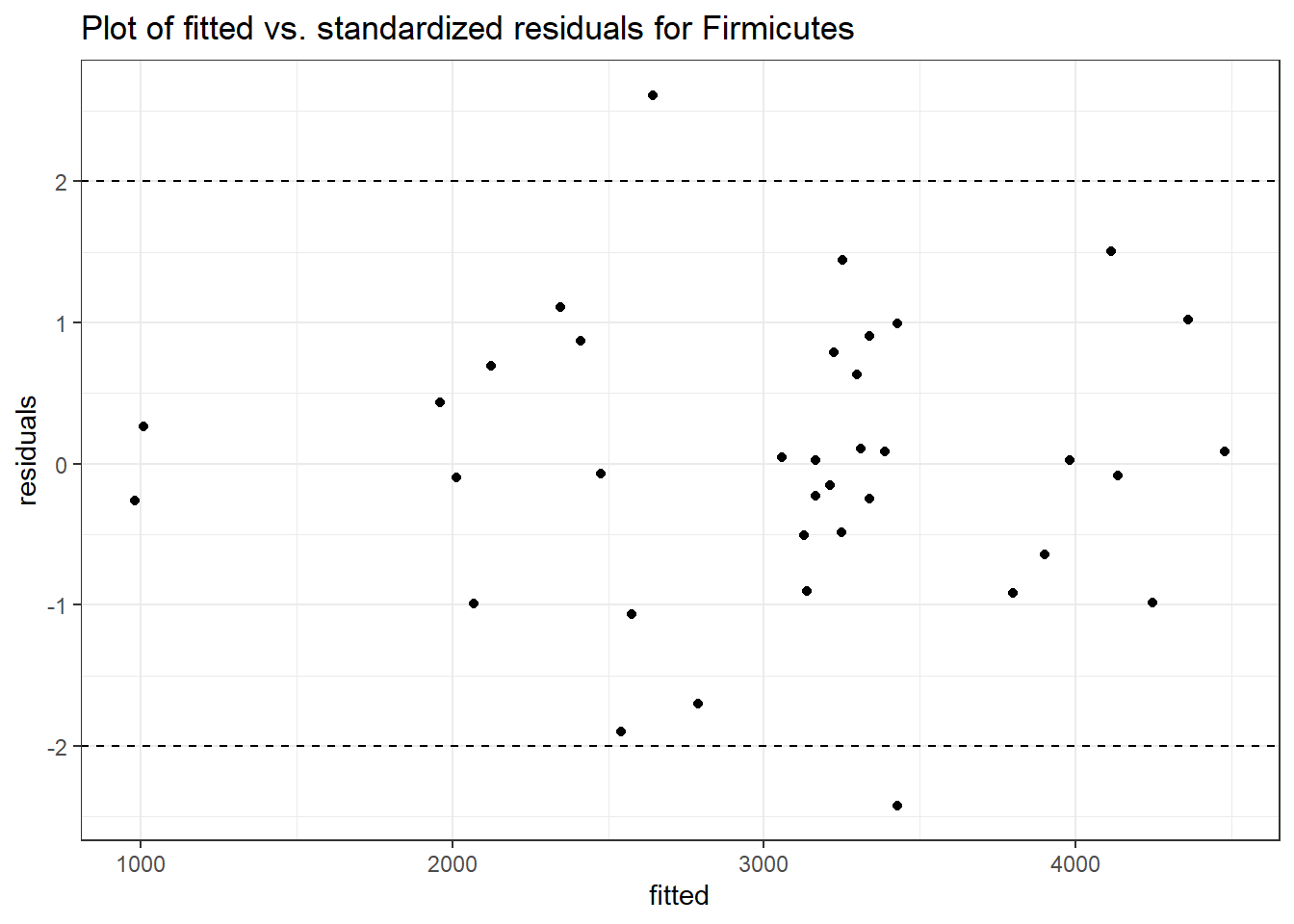

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.92162 0.11788 67.19813 0
time Firmicutes 0.02657 0.00267 9.96805 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.39064
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.8556128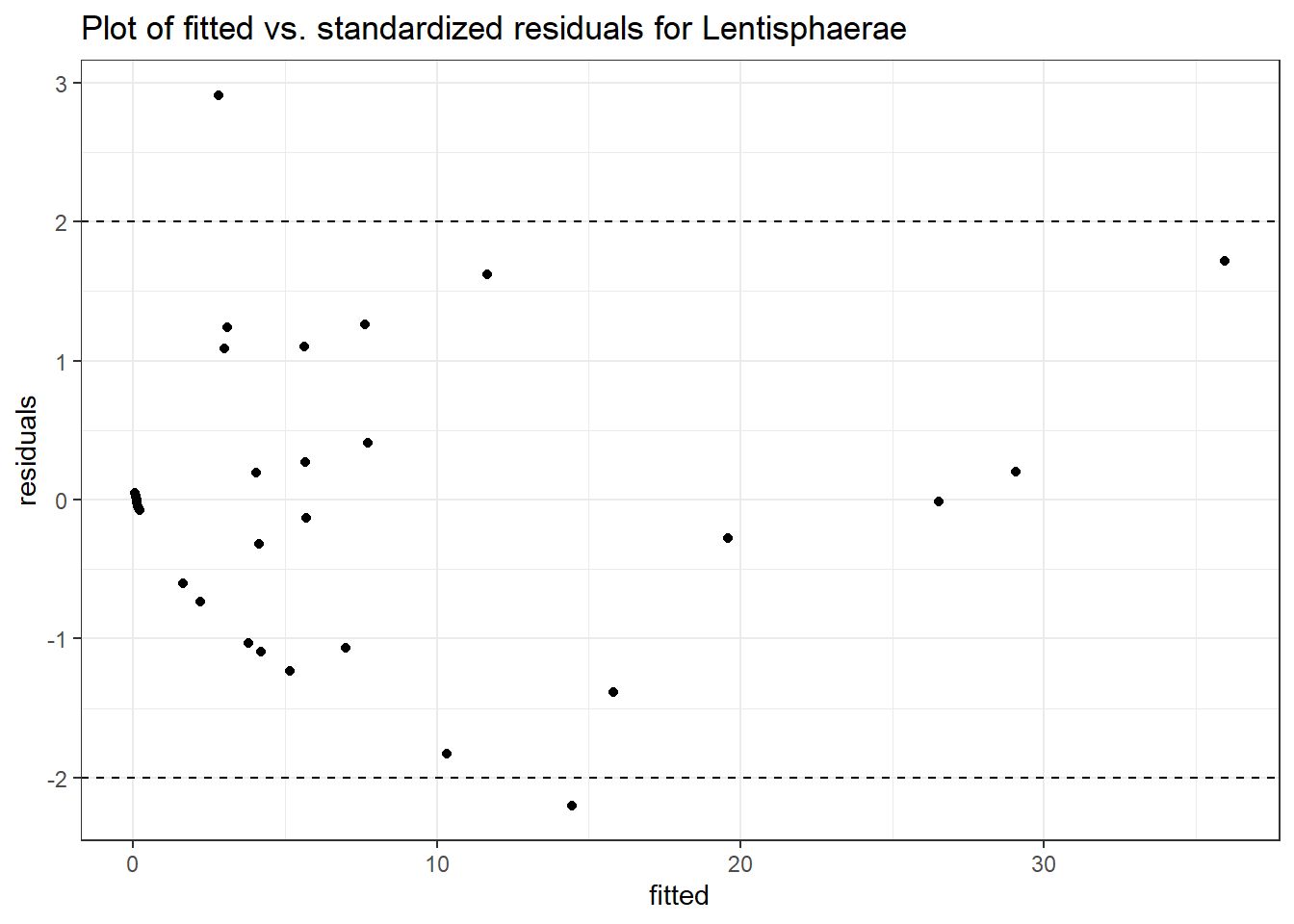

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae -0.15789 0.86771 -0.18197 0.85561
time Lentisphaerae 0.30395 0.05824 5.21848 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4343
Significance test not available for random effects
#########################################
#########################################
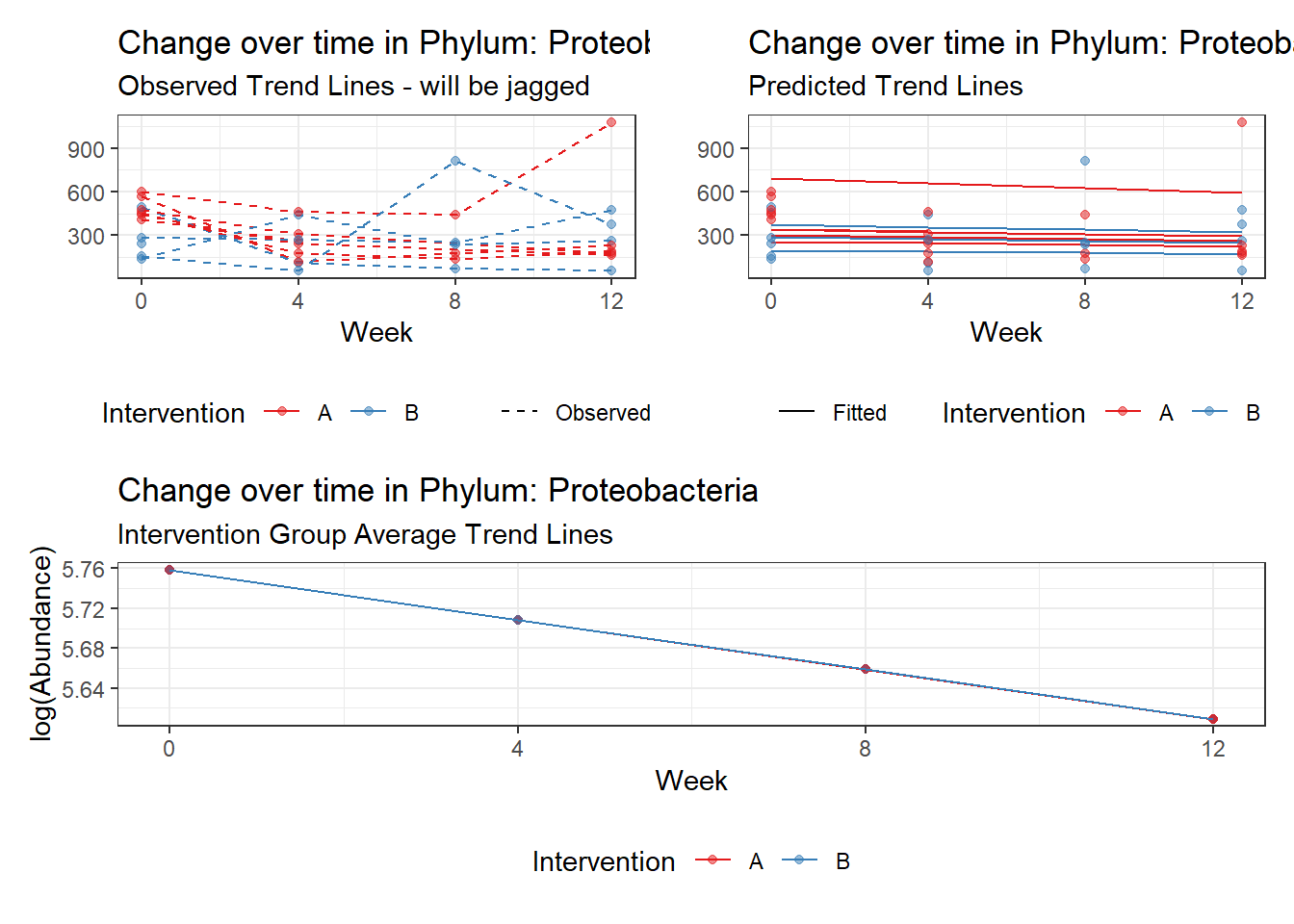
Model: Proteobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.08443468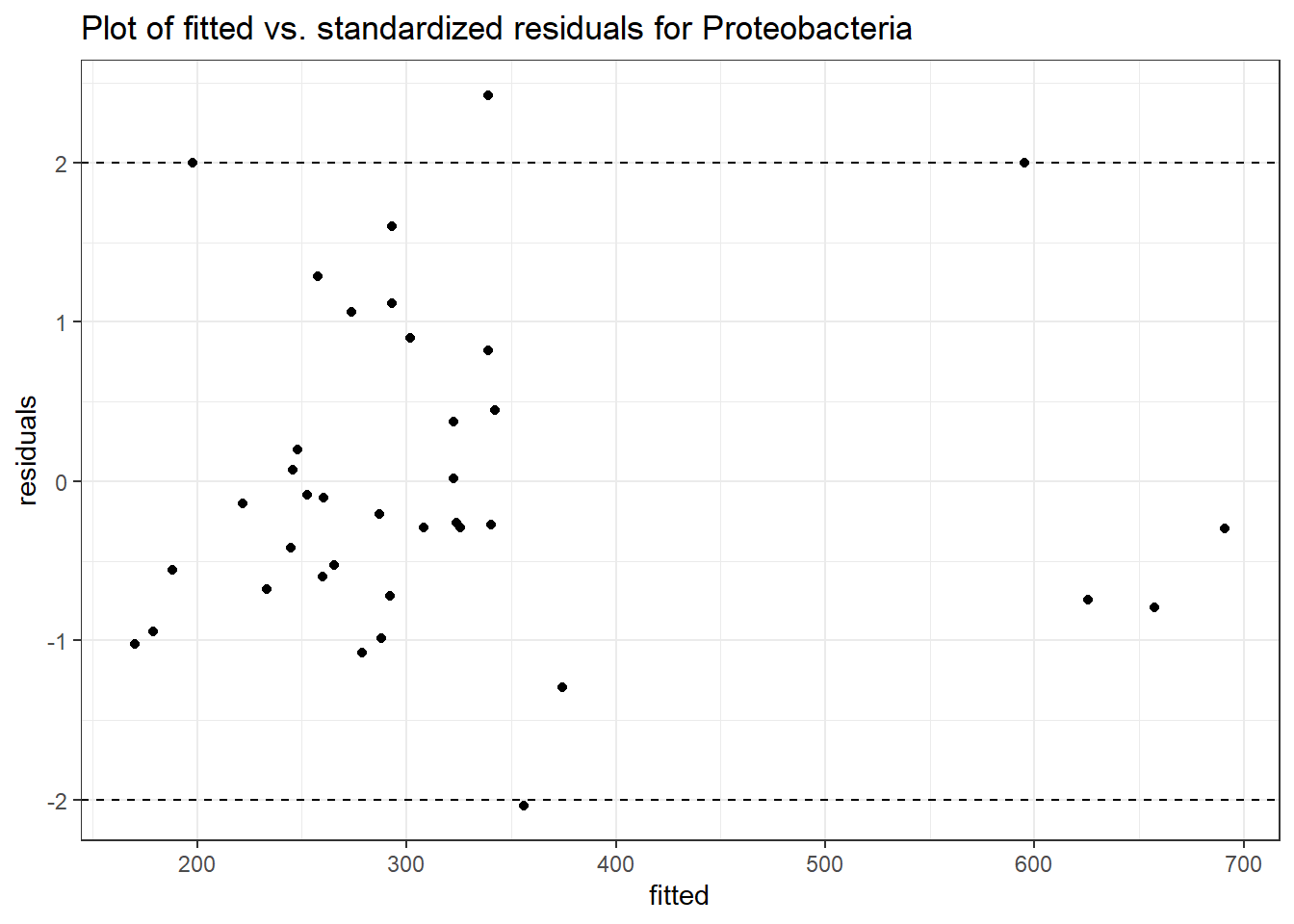
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.75835 0.09273 62.10061 0
time Proteobacteria -0.04969 0.00838 -5.93193 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.30368
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.9163098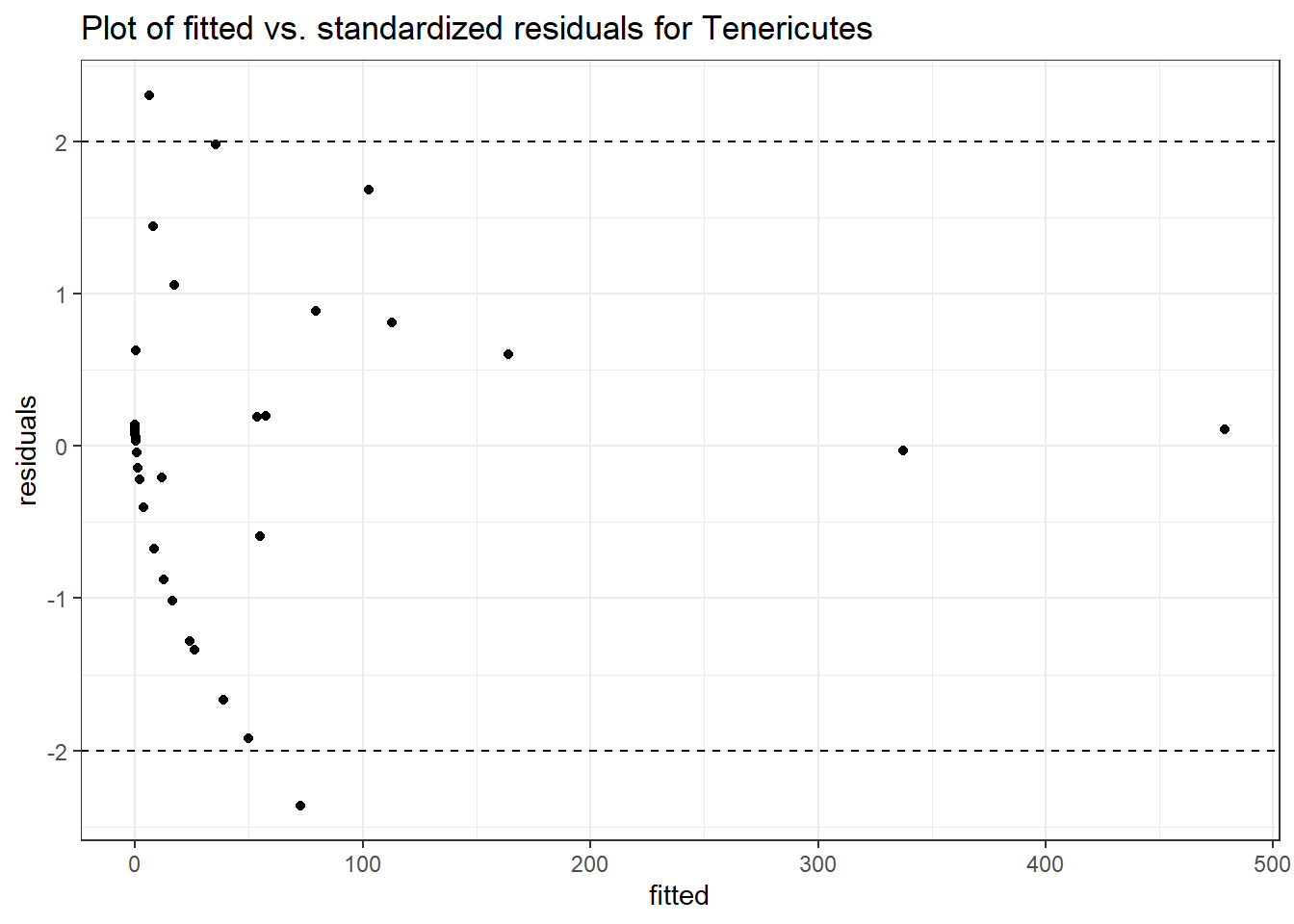
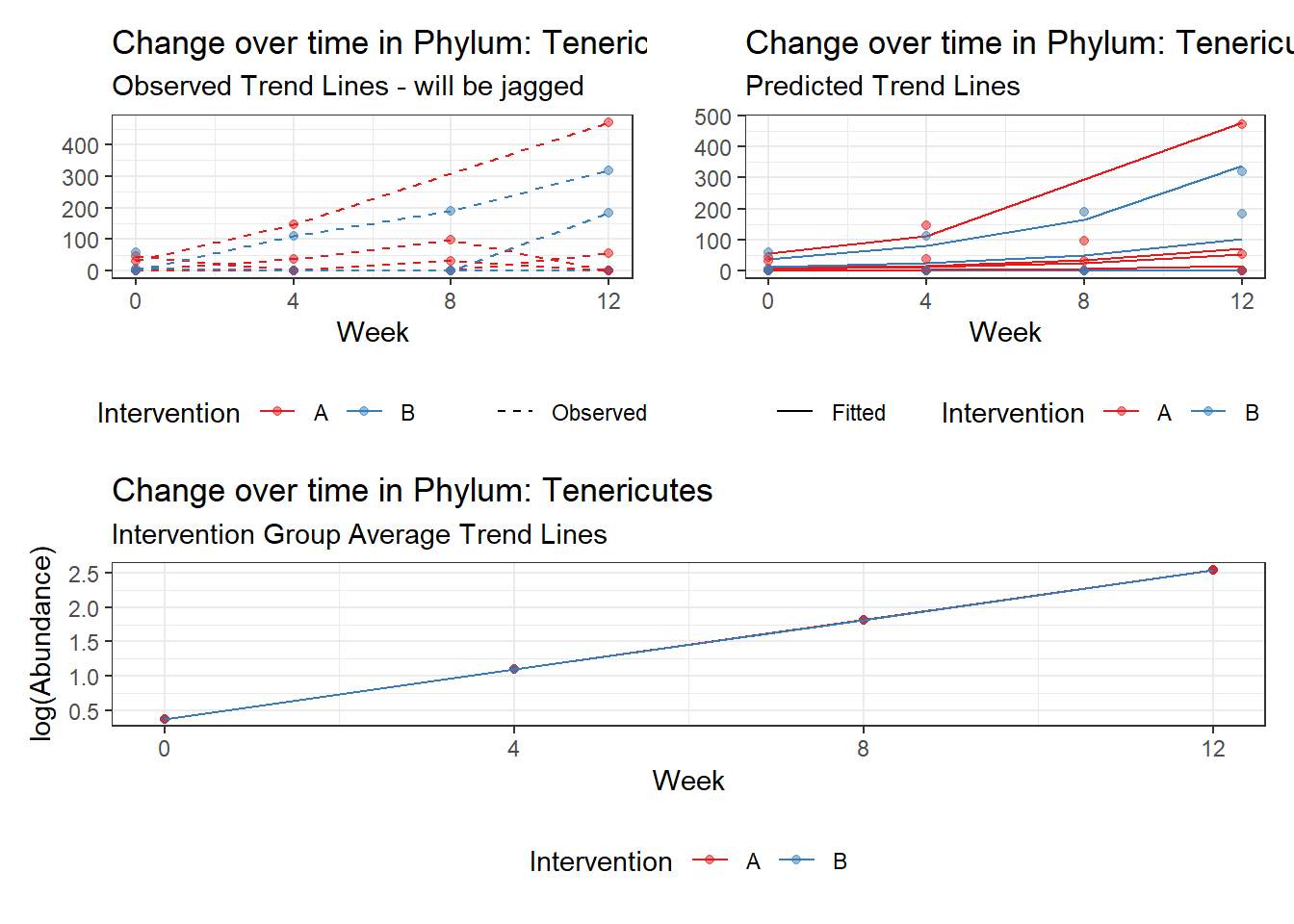
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 0.37537 1.03749 0.36181 0.71749
time Tenericutes 0.72193 0.02489 29.01006 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.3089
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000381f96c0>
Link: poisson
Intraclass Correlation (ICC): 0.8607546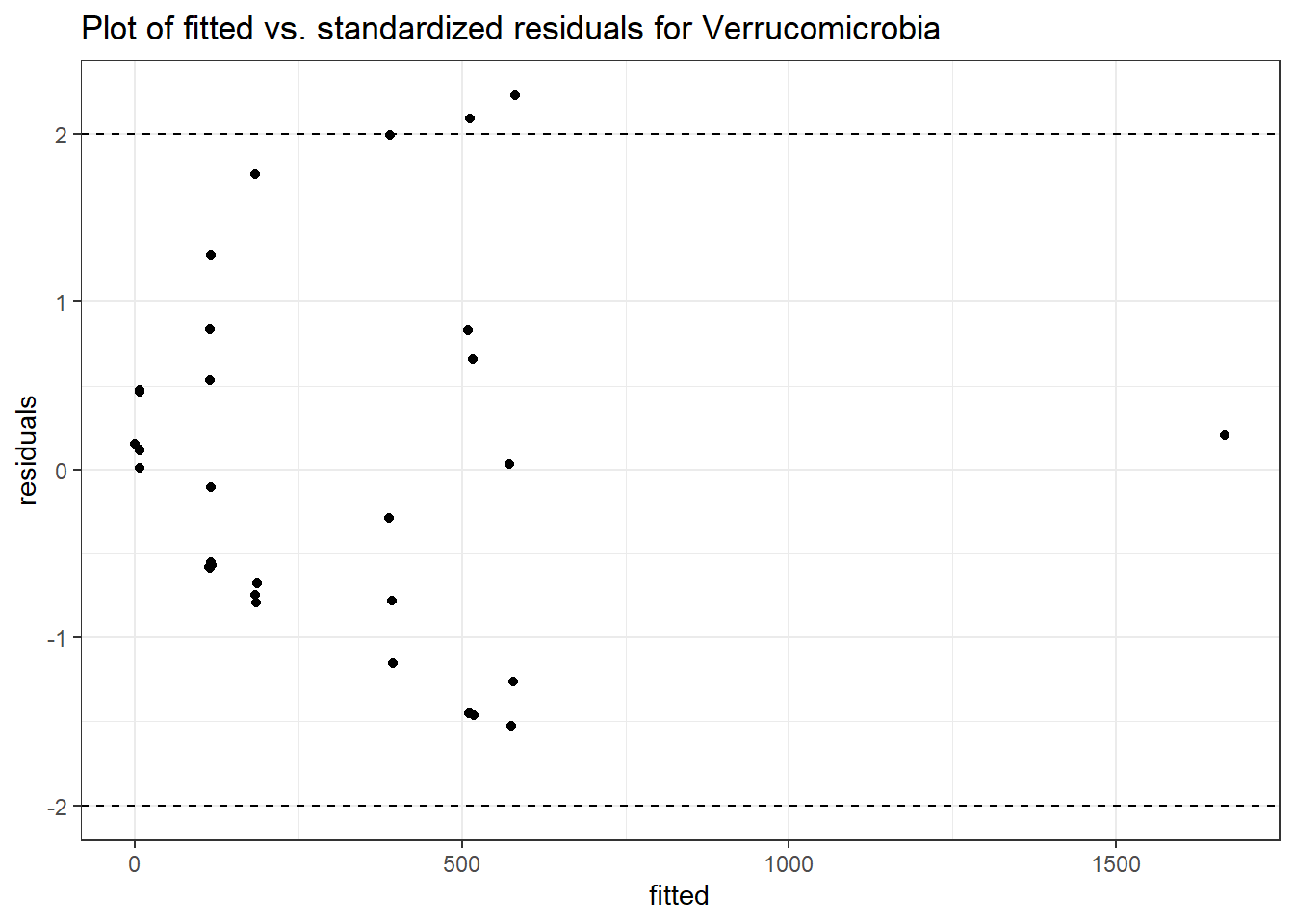
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.49846 0.75628 5.94816 0.00000
time Verrucomicrobia 0.00500 0.00920 0.54317 0.58701
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4863
Significance test not available for random effectsphylum.fit1b <- glmm_microbiome(mydata=microbiome_data,model.number=1,
taxa.level="Phylum", link="negbinom",
model="1 + time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinom
Intraclass Correlation (ICC): 0.375019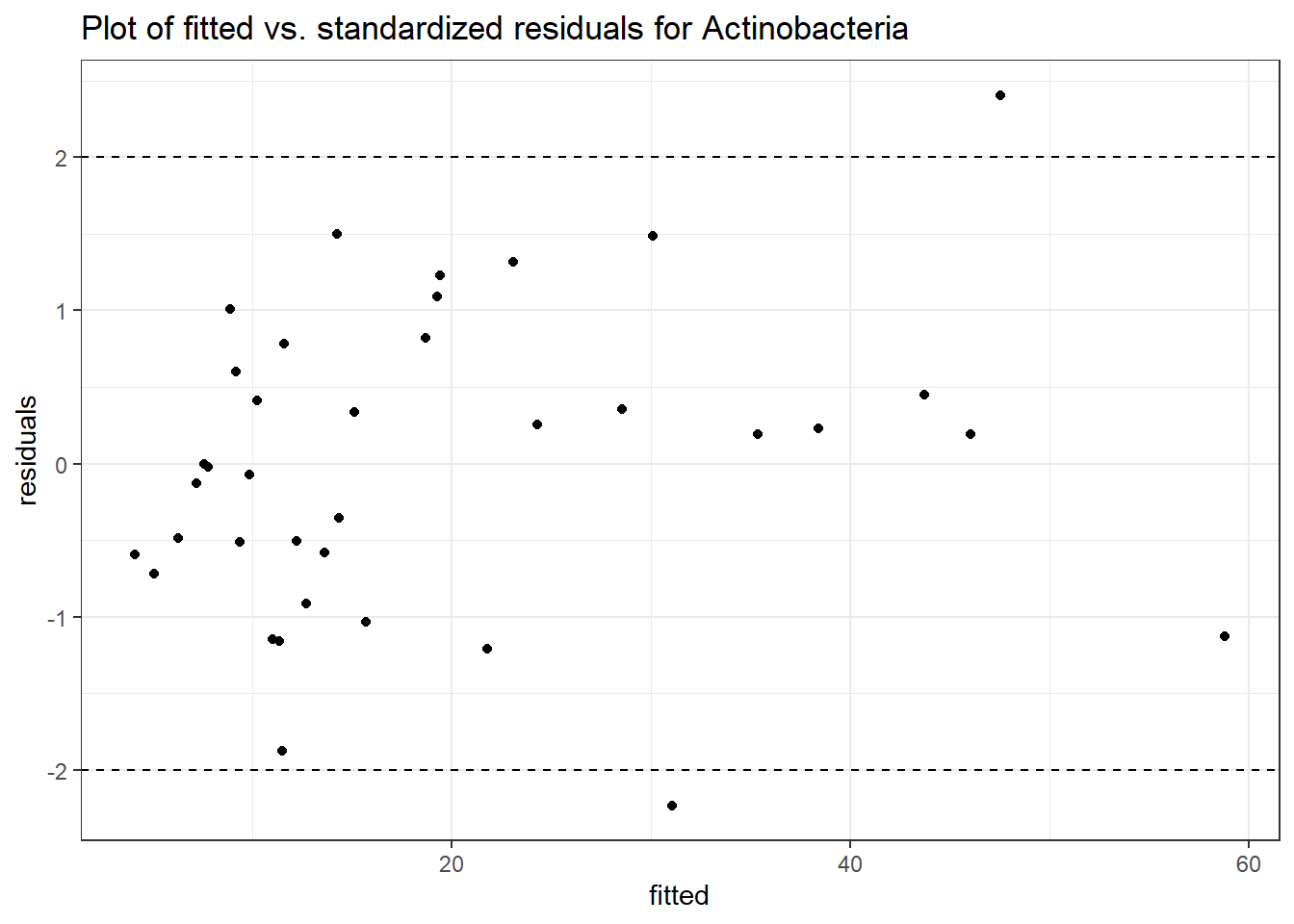
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.48613 0.40363 6.15948 0.00000
time Actinobacteria 0.21298 0.18887 1.12762 0.25948
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.77463
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinom
Intraclass Correlation (ICC): 0.142895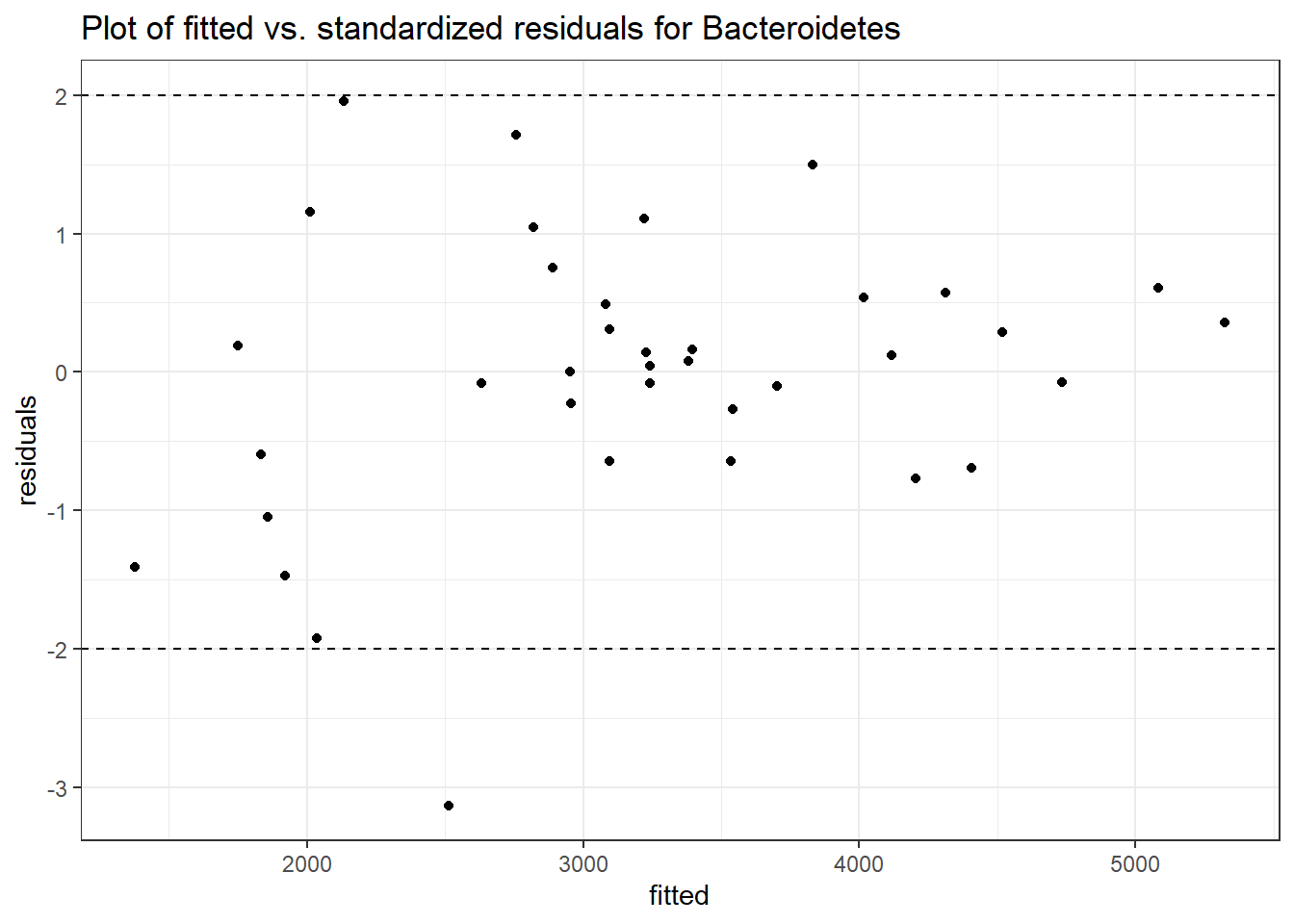
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.04573 0.13769 58.43425 0.00000
time Bacteroidetes -0.04644 0.03485 -1.33276 0.18261
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.40831
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 0
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 3.54247 1.27688 2.77432 0.00553
time Cyanobacteria -0.33819 0.79054 -0.42780 0.66880
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 3.497995e-10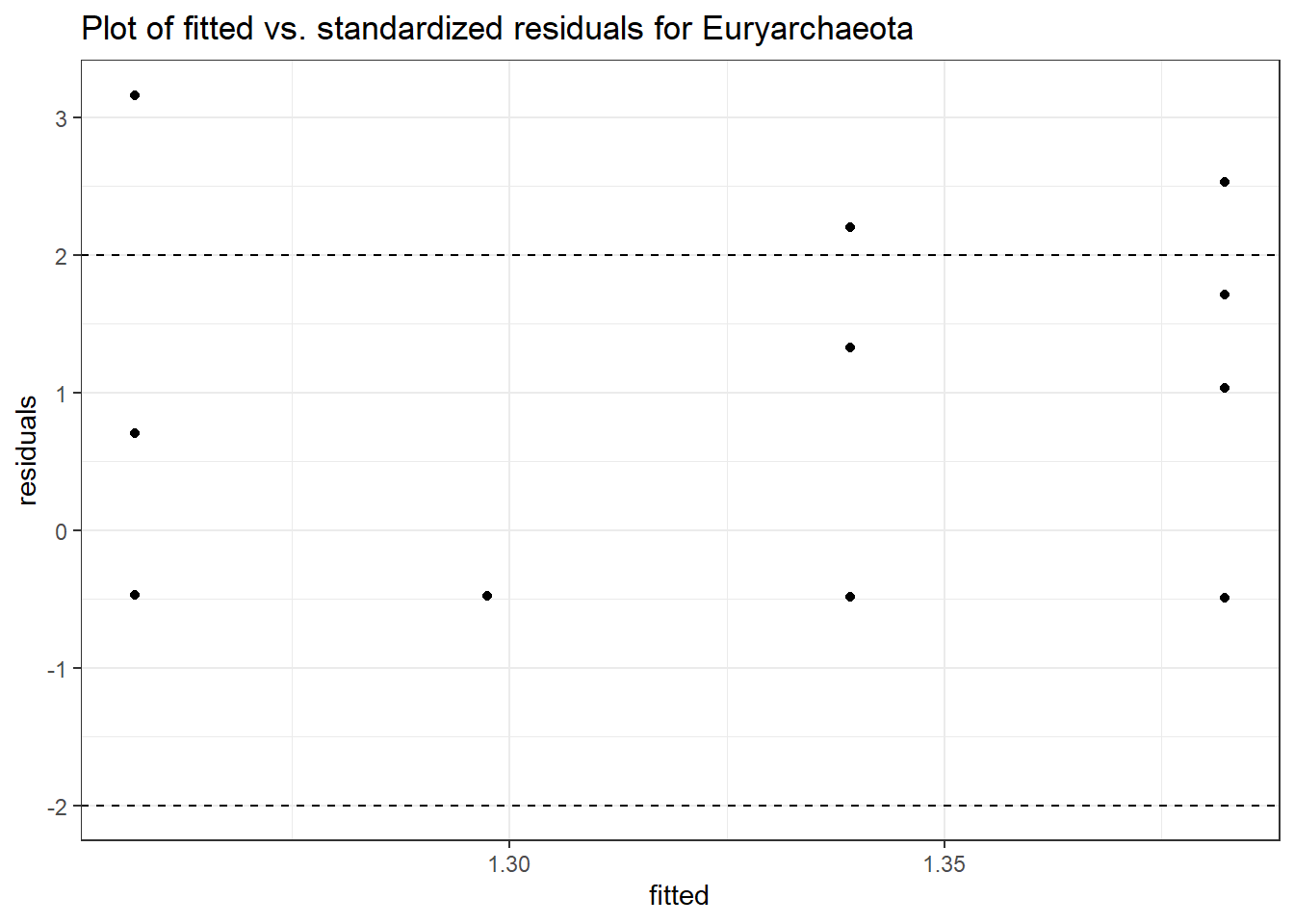
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota 0.32375 0.82223 0.39374 0.69377
time Euryarchaeota -0.03168 0.43726 -0.07245 0.94224
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.8703e-05
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinom
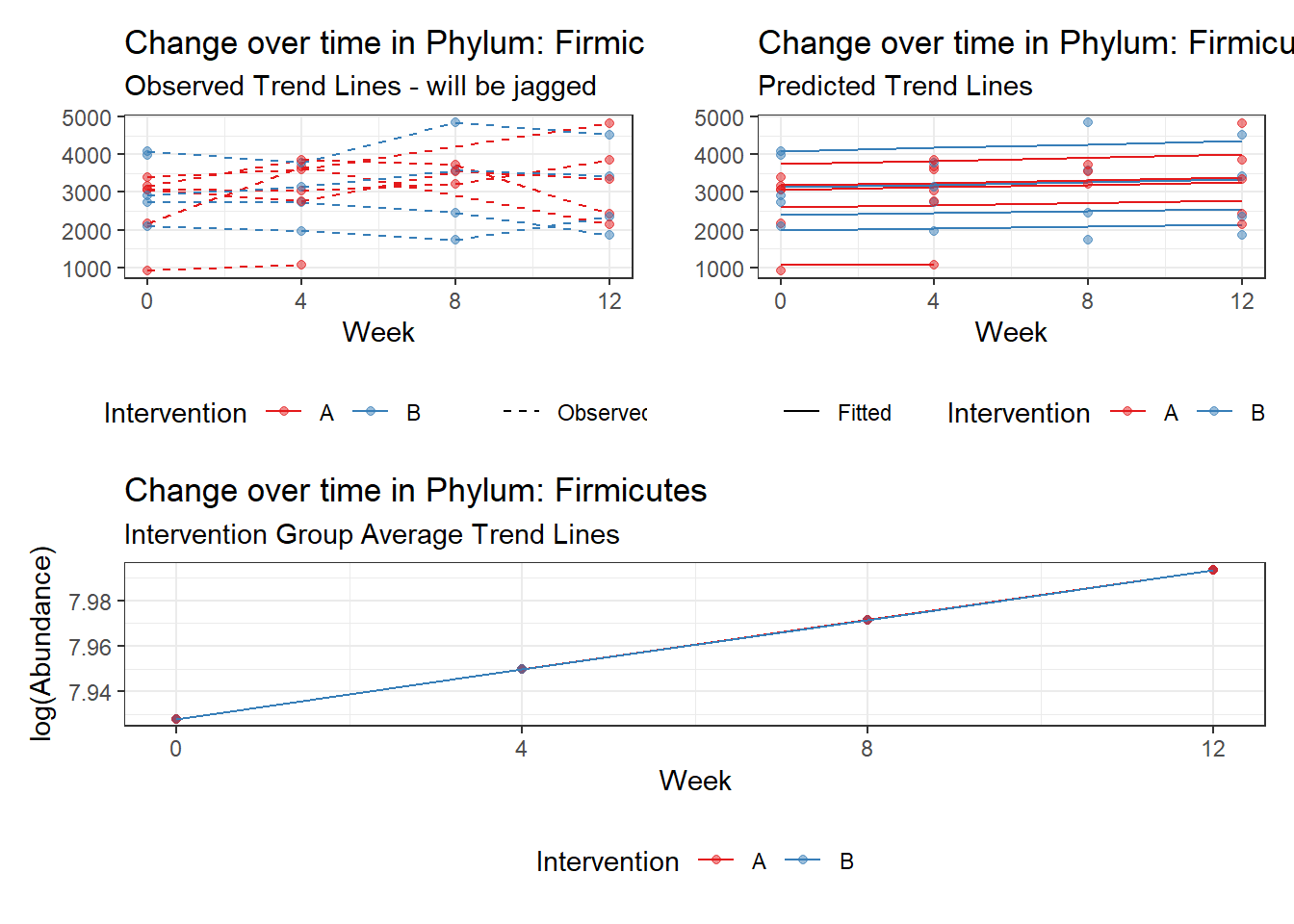
Intraclass Correlation (ICC): 0.1207101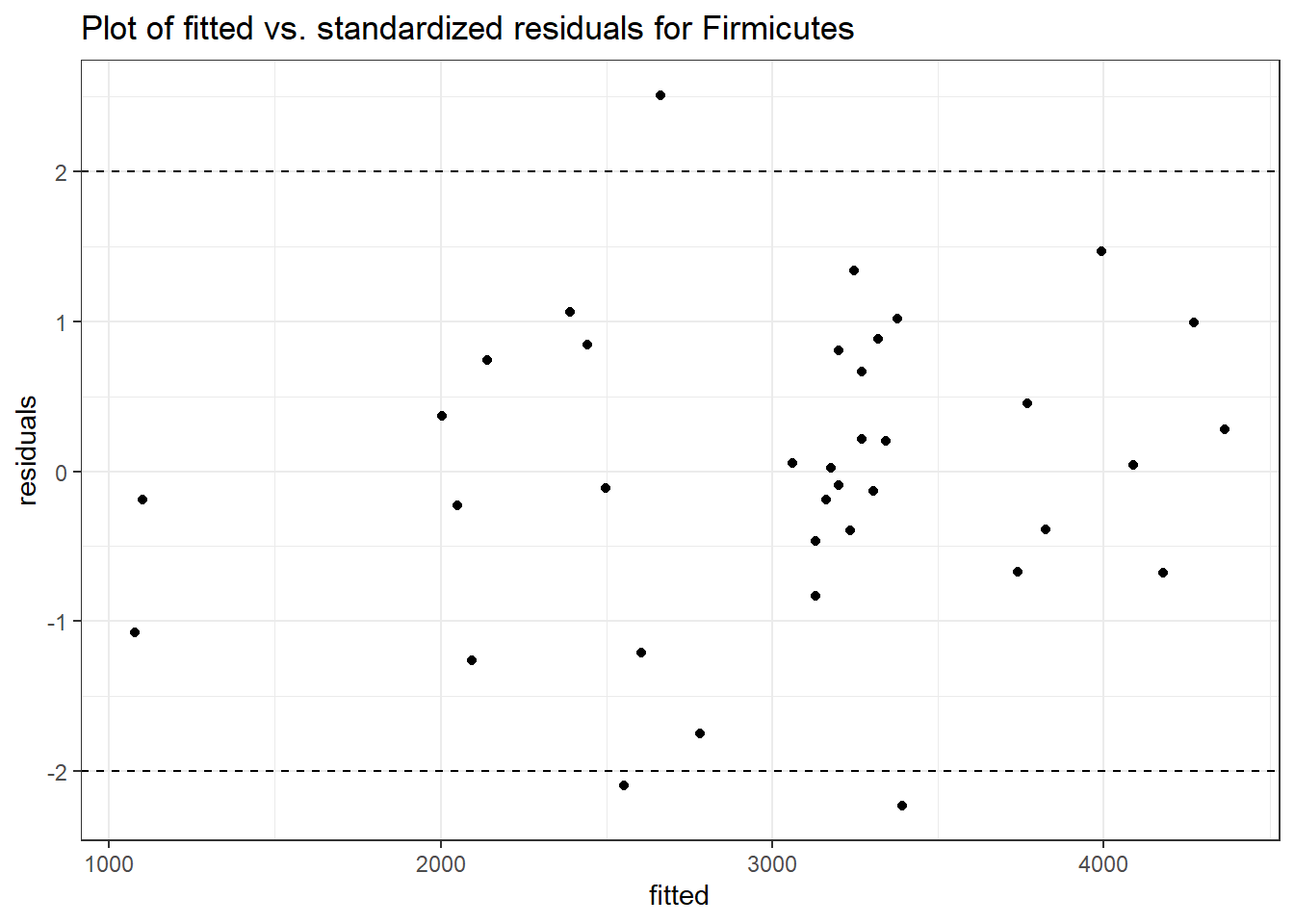
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.92796 0.11921 66.50188 0.00000
time Firmicutes 0.02184 0.02410 0.90630 0.36477
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.37051
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinom
Intraclass Correlation (ICC): 0.8168256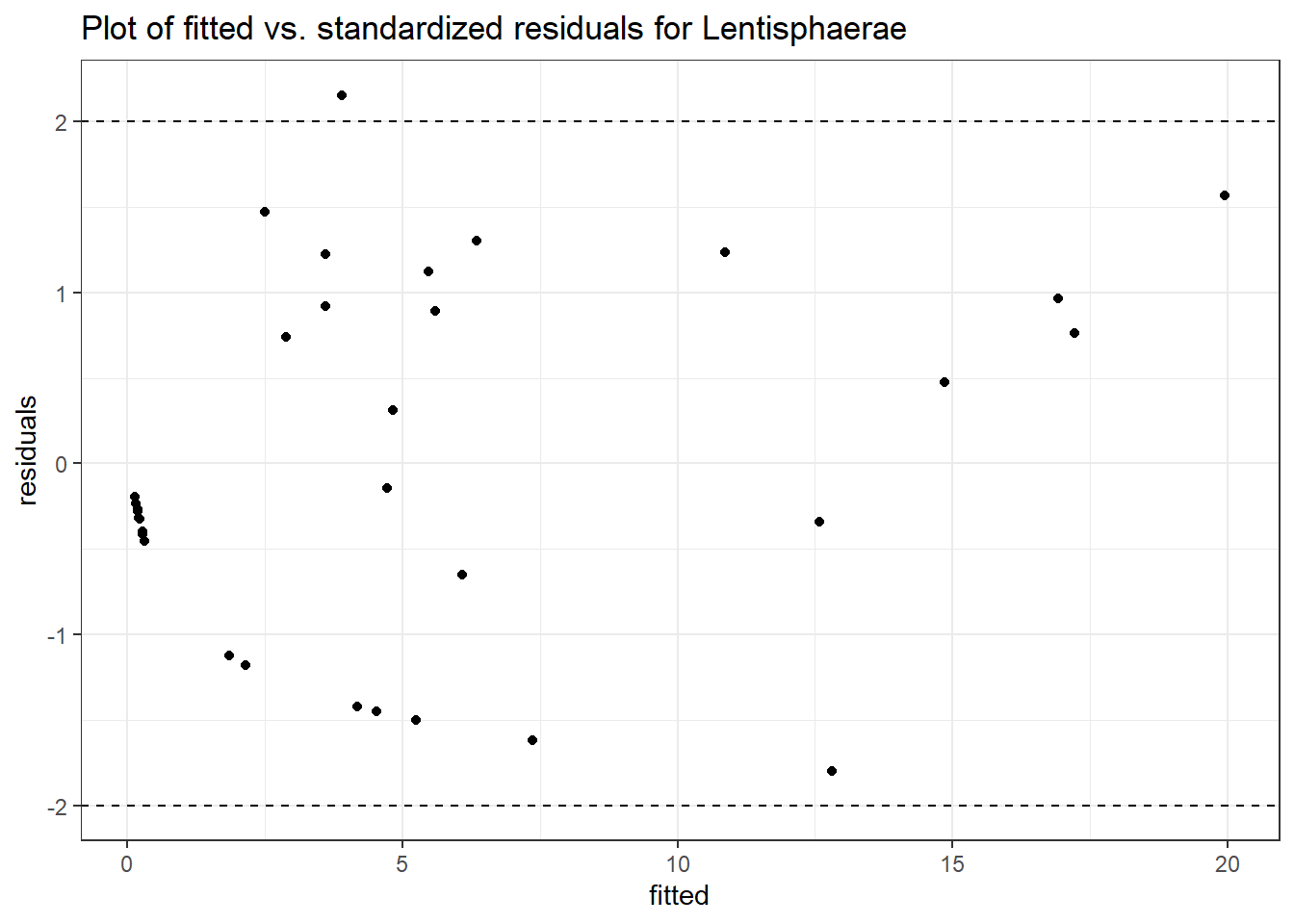

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.20406 0.92058 0.22166 0.82458
time Lentisphaerae 0.14781 0.28245 0.52331 0.60076
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1117
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinom
Intraclass Correlation (ICC): 0.03630509
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.81329 0.16269 35.73128 0.00000
time Proteobacteria -0.06285 0.08835 -0.71142 0.47683
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.19409
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinomboundary (singular) fit: see ?isSingular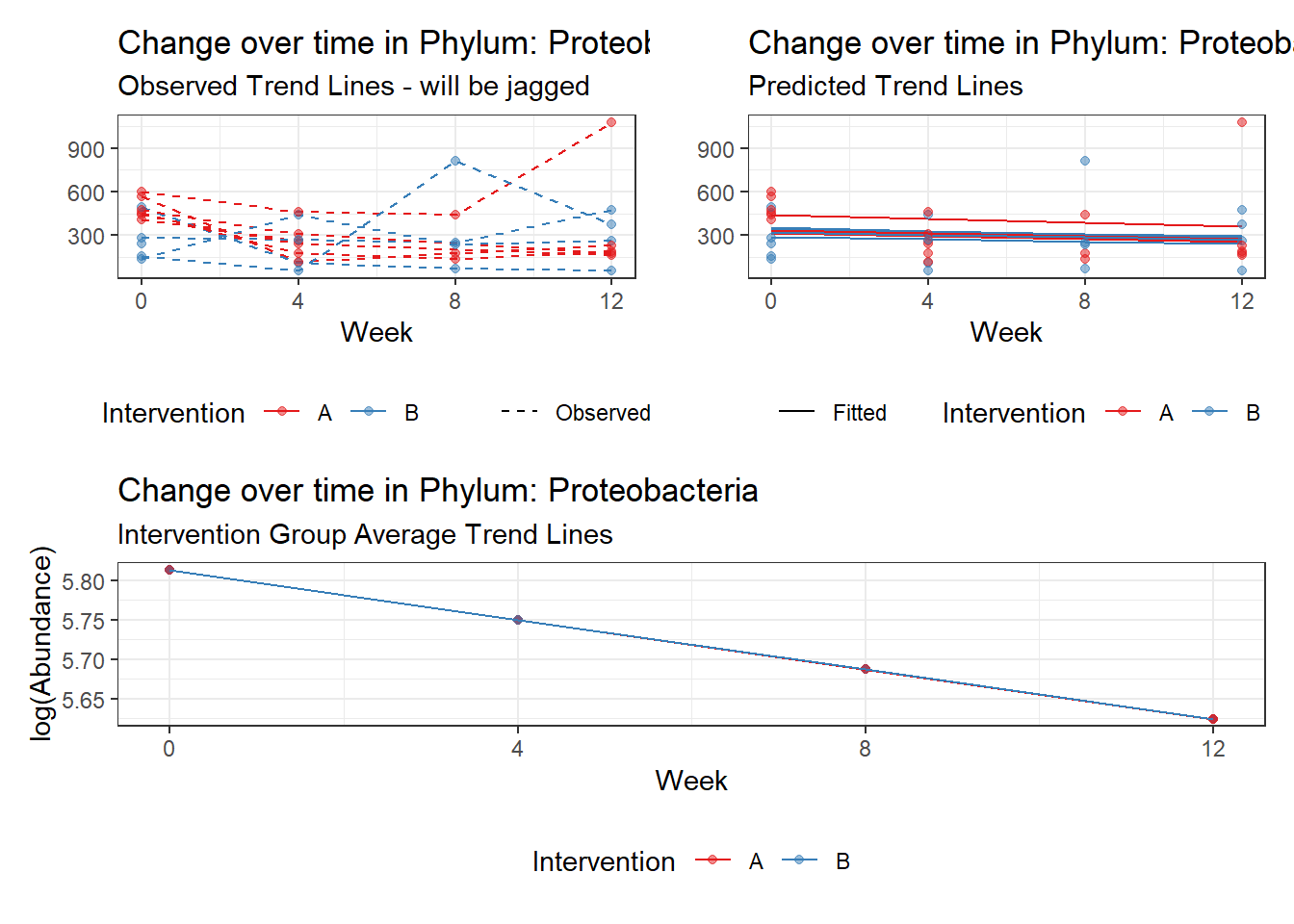
Intraclass Correlation (ICC): 0
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 2.57197 0.81255 3.16532 0.00155
time Tenericutes 0.70821 0.45254 1.56497 0.11759
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + (1 | SubjectID)
<environment: 0x000000003e389128>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced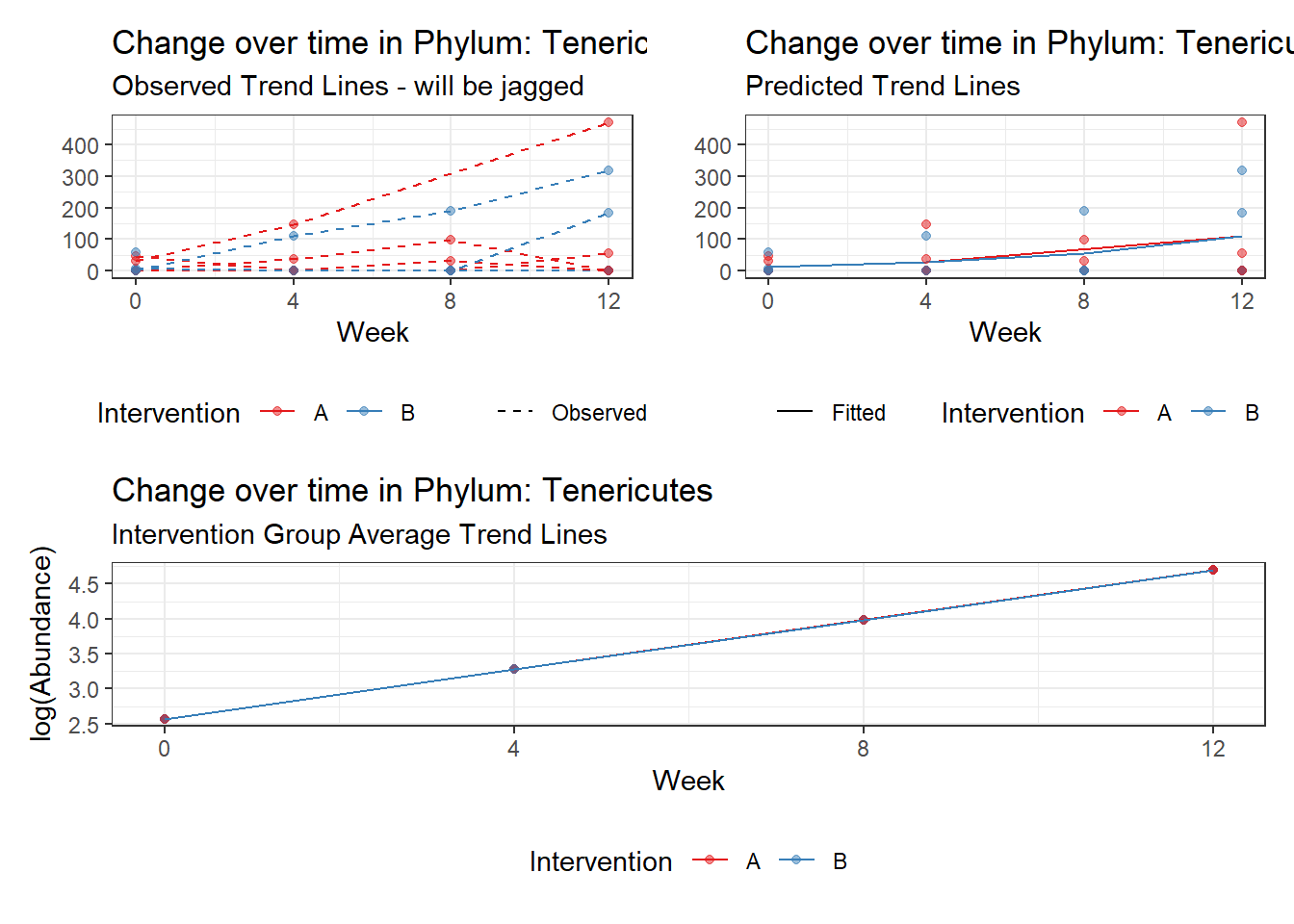
Intraclass Correlation (ICC): 0.8611511

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.50005 0.75659 5.94780 0.00000
time Verrucomicrobia 0.00196 0.00938 0.20909 0.83438
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4904
Significance test not available for random effectsSo, we found that time does significantly influence at least some of the bacteria over time. This is contrary to the results when we used the normal theory model.
Covariates: Time + Gender
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Gender)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
Gender is coded as female (effect of being female compared to males).
phylum.fit2 <- glmm_microbiome(mydata=microbiome_data,model.number=2,
taxa.level="Phylum", link="poisson",
model="1 + time + female + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poisson
Intraclass Correlation (ICC): 0.4982485
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.47700 0.50815 4.87455 0.00000
time Actinobacteria 0.17905 0.03052 5.86631 0.00000
female Actinobacteria 0.04105 0.63420 0.06472 0.94839
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.9965
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poisson
Intraclass Correlation (ICC): 0.1057896
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.43283 0.17206 49.01120 0.00000
time Bacteroidetes -0.03228 0.00265 -12.16667 0.00000
female Bacteroidetes -0.66491 0.21571 -3.08251 0.00205
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.34395
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poisson
Intraclass Correlation (ICC): 0.9488445

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -3.69881 3.11001 -1.18932 0.23431
time Cyanobacteria -0.27256 0.03201 -8.51544 0.00000
female Cyanobacteria 3.96040 3.29269 1.20278 0.22906
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.3068
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poisson
Intraclass Correlation (ICC): 0.7603834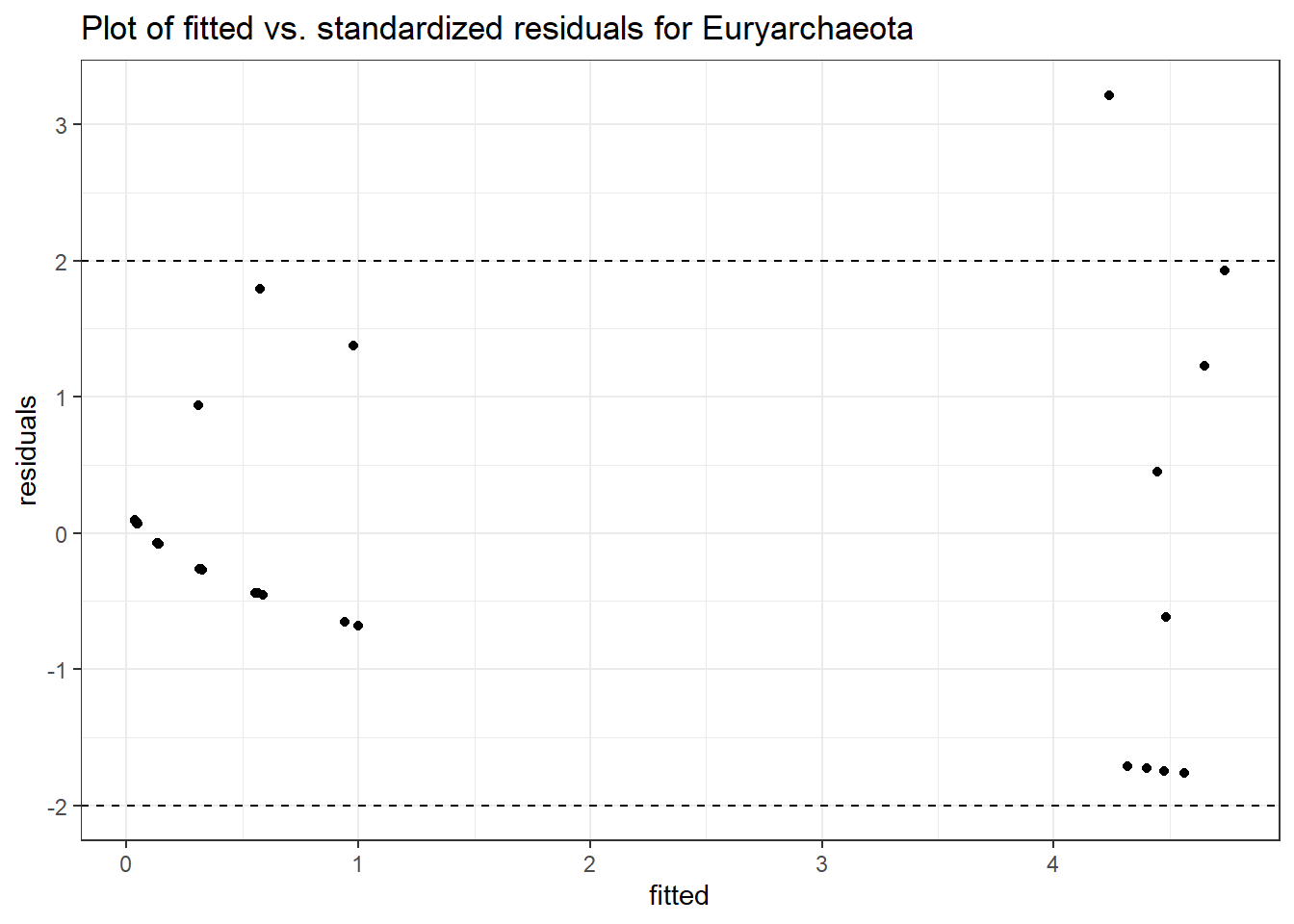
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -2.78911 1.45323 -1.91925 0.05495
time Euryarchaeota -0.01899 0.13261 -0.14320 0.88613
female Euryarchaeota 2.53223 1.52281 1.66287 0.09634
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.7814
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poisson
Intraclass Correlation (ICC): 0.05733286
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.52153 0.12357 60.86953 0e+00
time Firmicutes 0.02659 0.00267 9.97672 0e+00
female Firmicutes 0.62883 0.15479 4.06239 5e-05
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.24662
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poissonWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
unable to evaluate scaled gradientWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge: degenerate Hessian with 1 negative eigenvalues
Intraclass Correlation (ICC): 0.3388261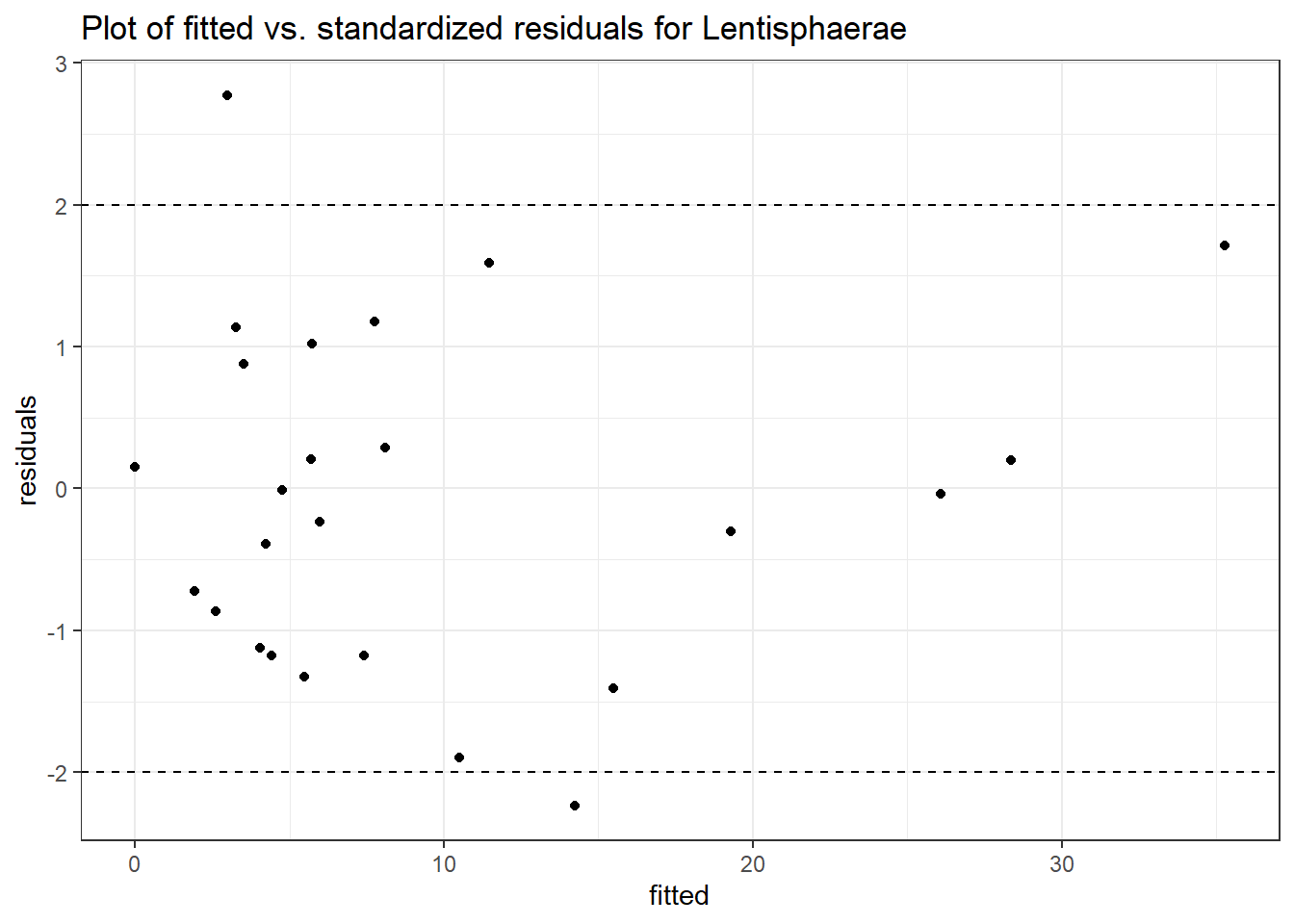
Warning in vcov.merMod(object, use.hessian = use.hessian): variance-covariance matrix computed from finite-difference Hessian is
not positive definite or contains NA values: falling back to var-cov estimated from RXWarning in vcov.merMod(object, correlation = correlation, sigm = sig): variance-covariance matrix computed from finite-difference Hessian is
not positive definite or contains NA values: falling back to var-cov estimated from RX
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae -18.83511 2716.82272 -0.00693 0.99447
time Lentisphaerae 0.30236 0.05792 5.22067 0.00000
female Lentisphaerae 20.41620 2716.82274 0.00751 0.99400
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.71586
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poisson
Intraclass Correlation (ICC): 0.08001611
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.66360 0.14879 38.06363 0.00000
time Proteobacteria -0.04970 0.00838 -5.93280 0.00000
female Proteobacteria 0.14908 0.18616 0.80083 0.42323
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.29492
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poisson
Intraclass Correlation (ICC): 0.5764745
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes -3.79522 1.06416 -3.56640 0.00036
time Tenericutes 0.72079 0.02488 28.97019 0.00000
female Tenericutes 6.47823 1.15036 5.63146 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.1667
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000003a582e10>
Link: poisson
Intraclass Correlation (ICC): 0.827486
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 2.90035 1.12841 2.57031 0.01016
time Verrucomicrobia 0.00500 0.00920 0.54291 0.58719
female Verrucomicrobia 2.50316 1.39938 1.78876 0.07365
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1901
Significance test not available for random effectsphylum.fit2b <- glmm_microbiome(mydata=microbiome_data,model.number=2,
taxa.level="Phylum", link="negbinom",
model="1 + time + female + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046721990>
Link: negbinom
Intraclass Correlation (ICC): 0.3728722
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.55542 0.57905 4.41310 0.00001
time Actinobacteria 0.21316 0.18843 1.13125 0.25795
female Actinobacteria -0.10645 0.64228 -0.16574 0.86836
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.77108
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + female + (1 | SubjectID)
<environment: 0x0000000046721990>
Link: negbinomError in factory(refitNB, types = c("message", "warning"))(lastfit, theta = exp(t), : Downdated VtV is not positive definite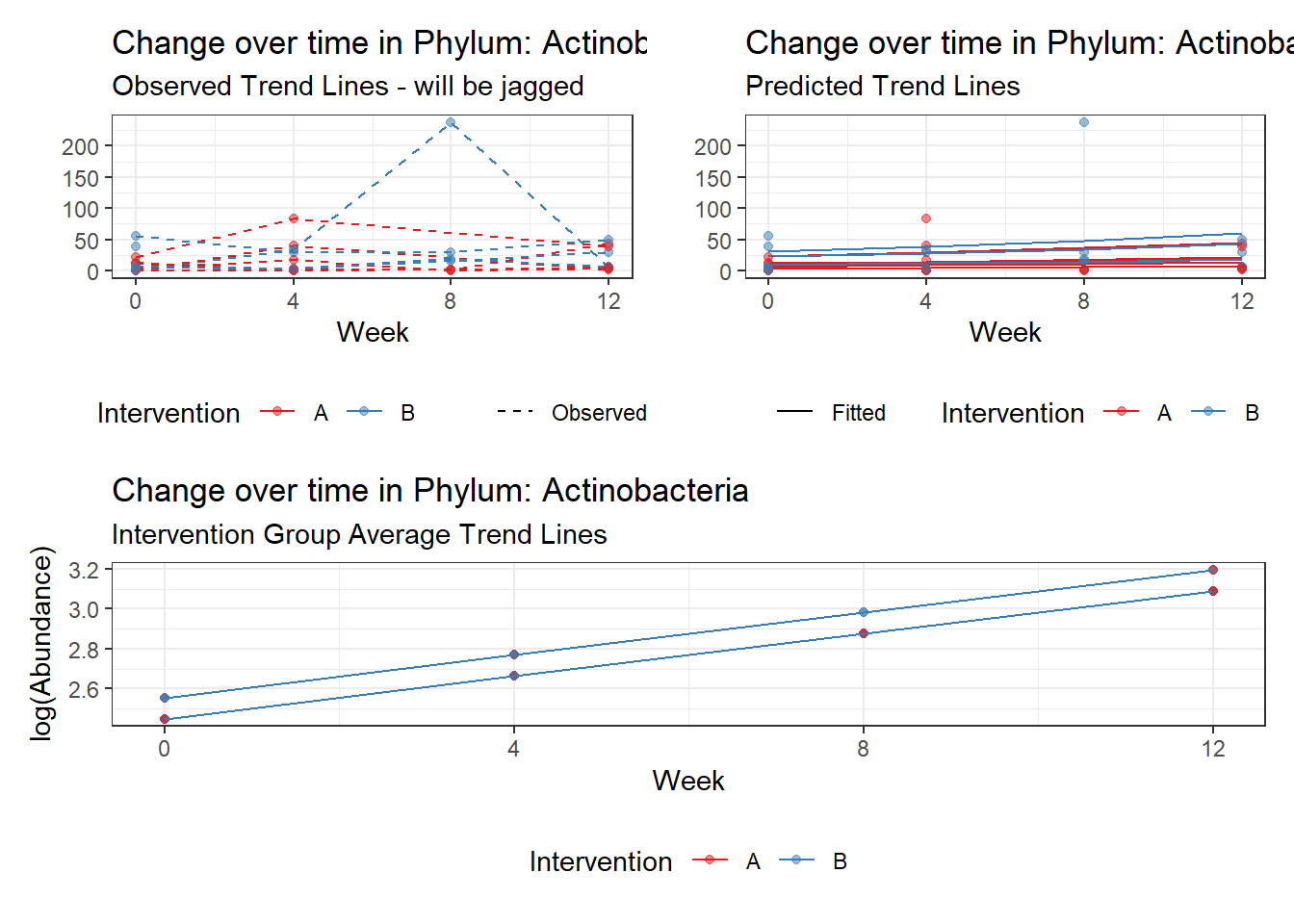
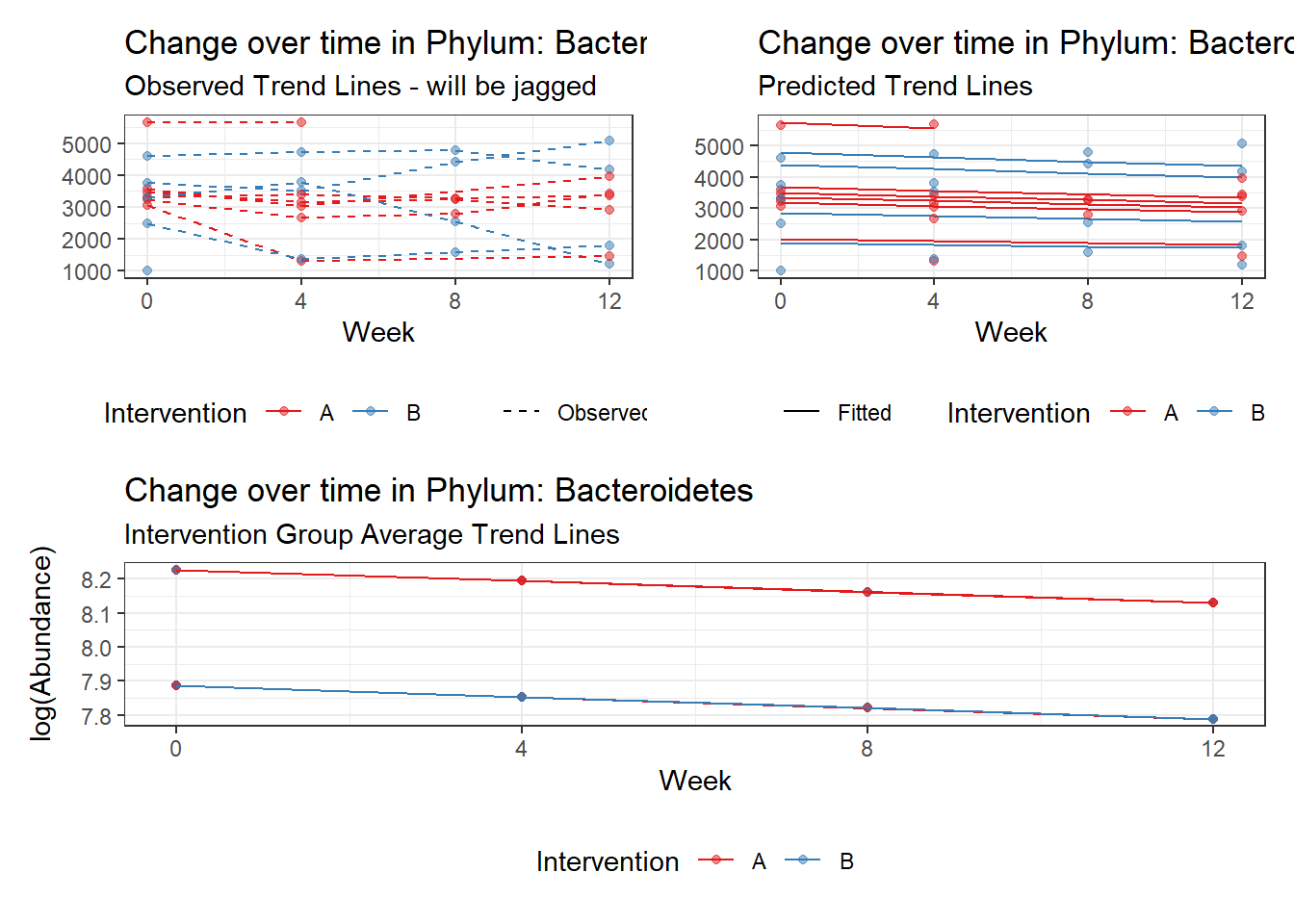
Controlling for the effect of time, Gender was only signficant for Bacteroidetes and Firmicutes. We will take out gender out of model to test other effects, but will come back when testing intervention.
Covariates: Time + Ethnicity
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Ethnicity)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
Ethnicity is coded as Hispanic (effect of being Hispanic compared to White, Asian or Native American). Groups were coded this way due to sample size.
phylum.fit3 <- glmm_microbiome(mydata=microbiome_data,model.number=3,
taxa.level="Phylum", link="poisson",
model="1 + time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.4798439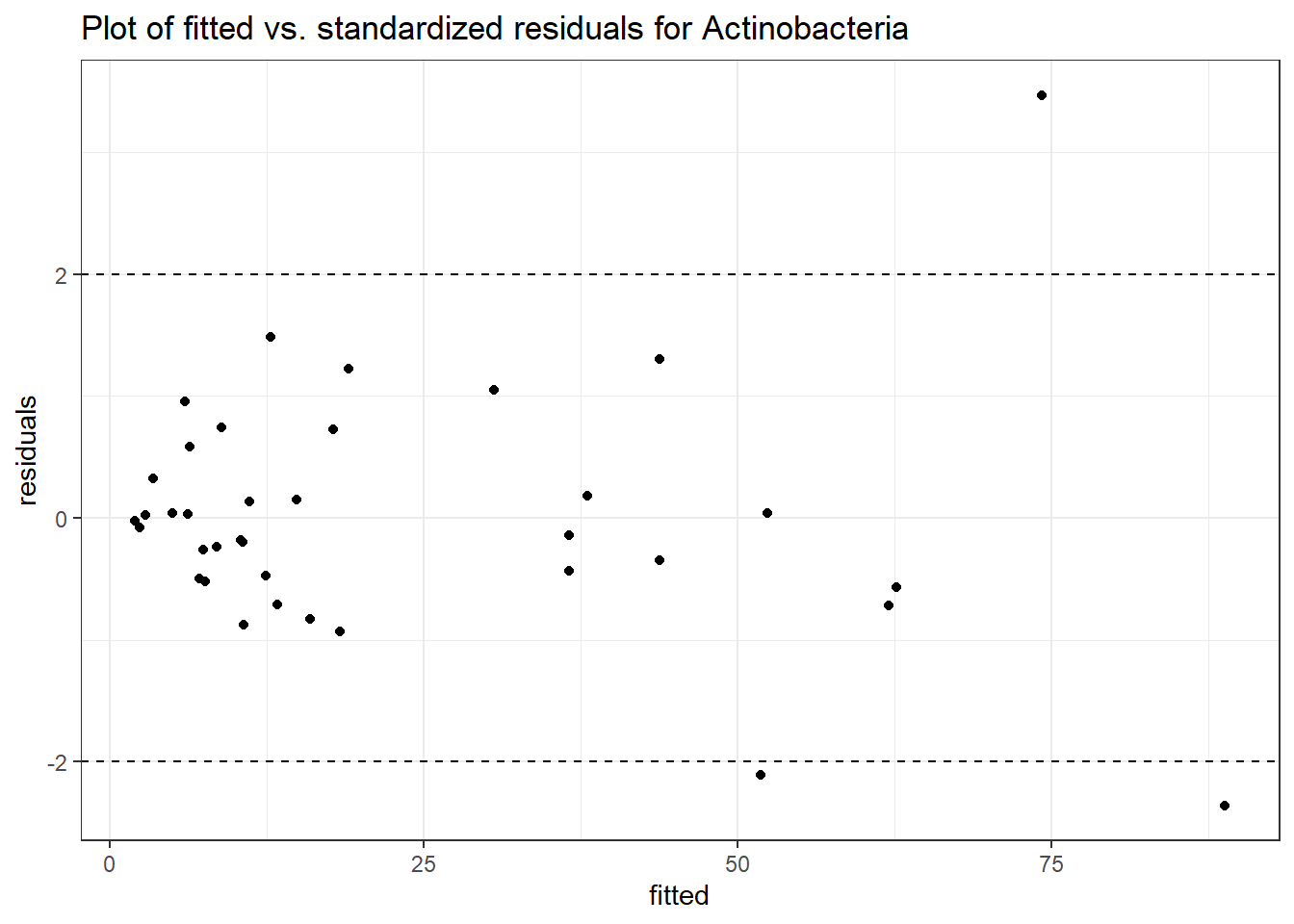
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.13350 0.49326 4.32533 0.00002
time Actinobacteria 0.17954 0.03053 5.88069 0.00000
hispanic Actinobacteria 0.57933 0.61313 0.94488 0.34472
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.96047
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.1625324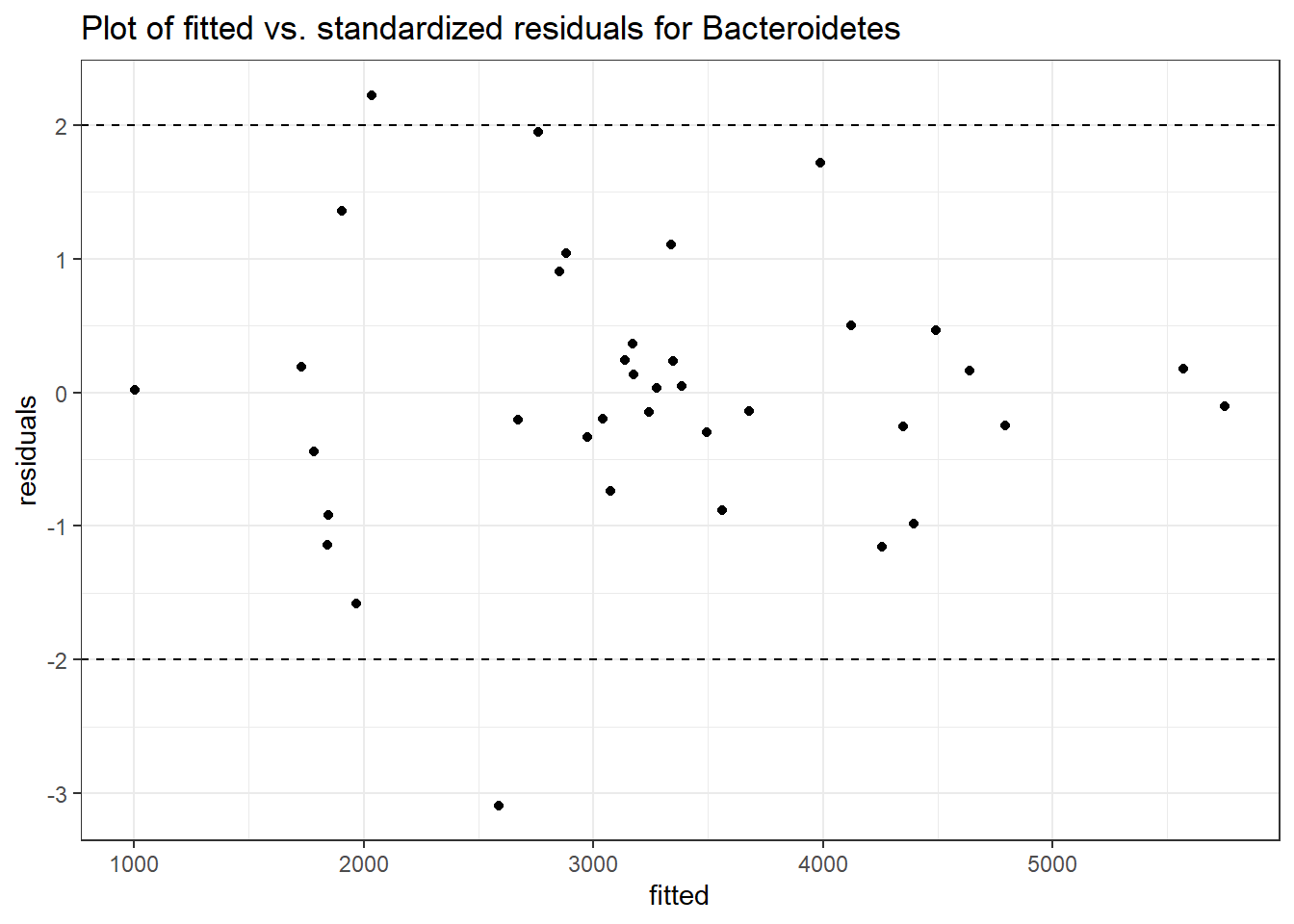
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.22674 0.22034 37.33580 0.00000
time Bacteroidetes -0.03232 0.00265 -12.18155 0.00000
hispanic Bacteroidetes -0.34125 0.27622 -1.23544 0.21667
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.44054
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.9381702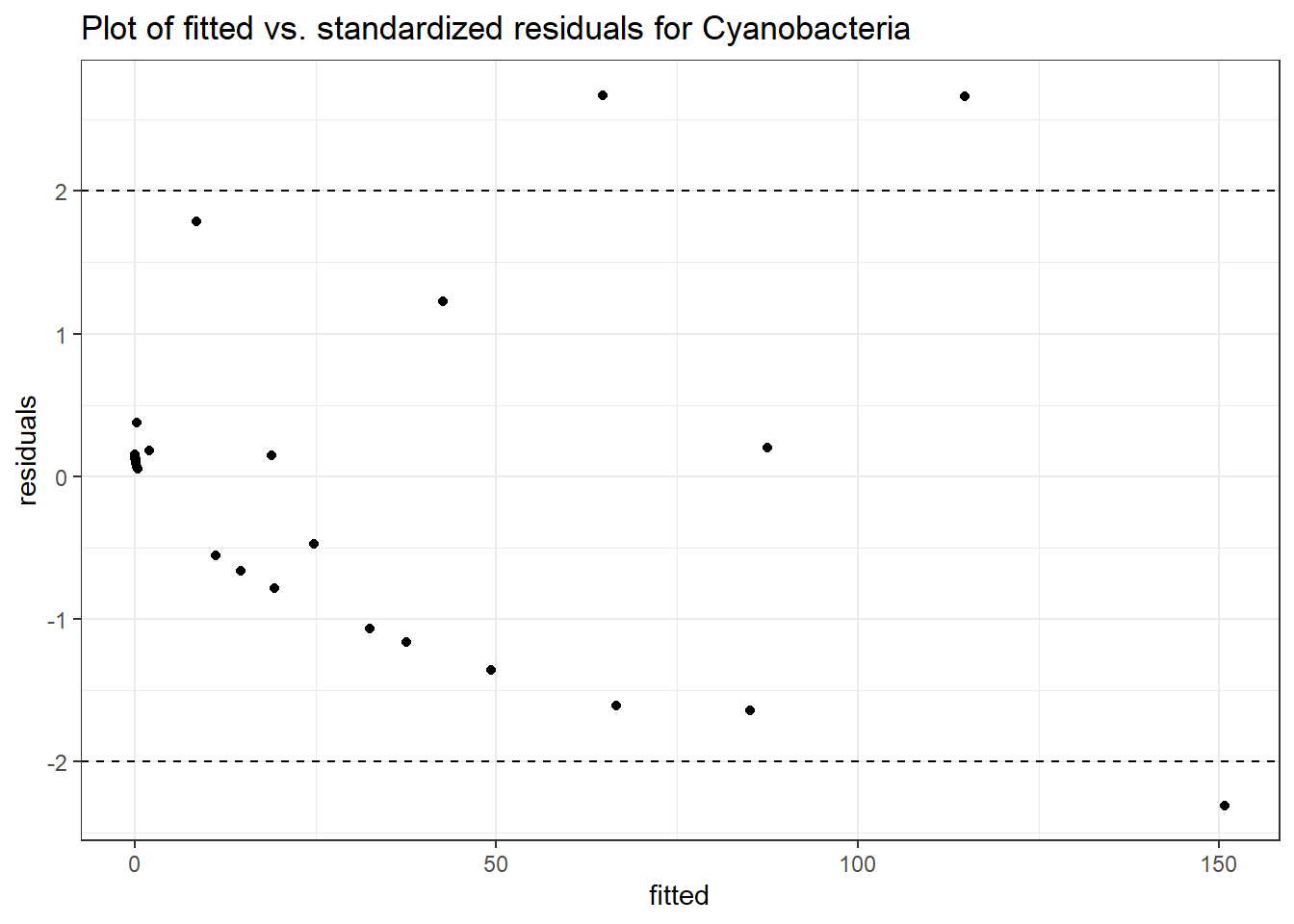
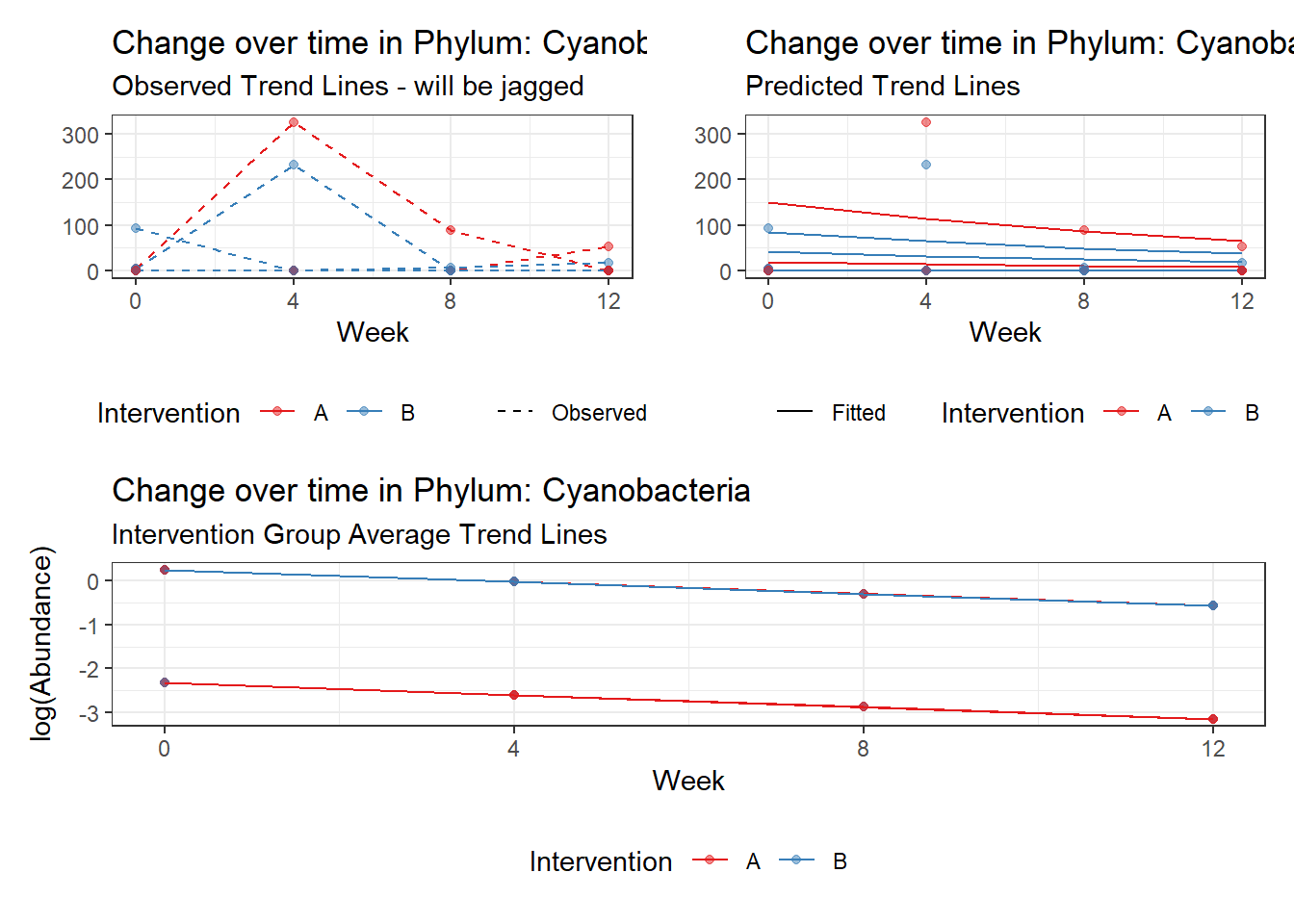
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -2.32857 2.48673 -0.93640 0.34907
time Cyanobacteria -0.27244 0.03201 -8.51175 0.00000
hispanic Cyanobacteria 2.57061 2.81681 0.91260 0.36145
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.8953
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.6356139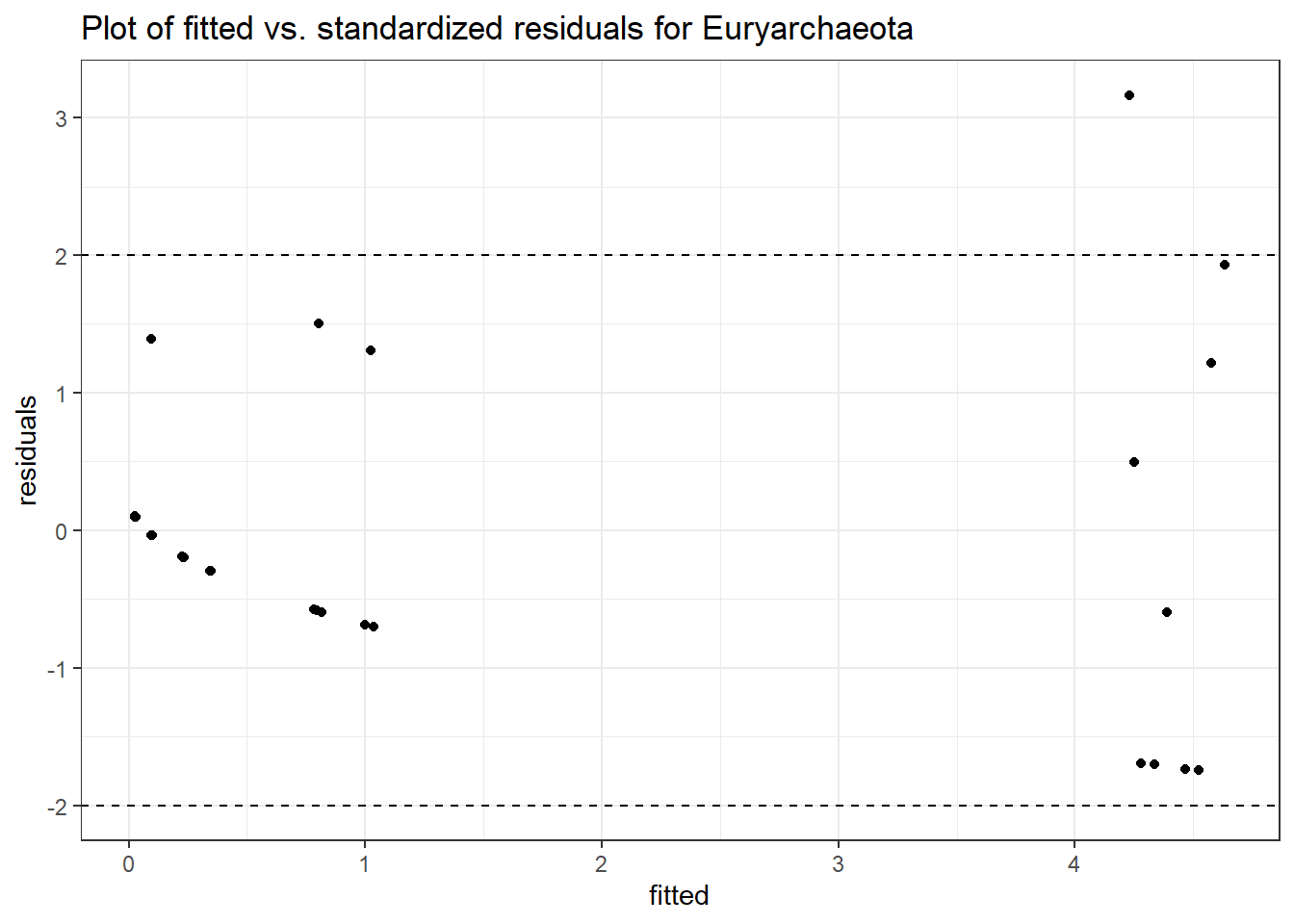
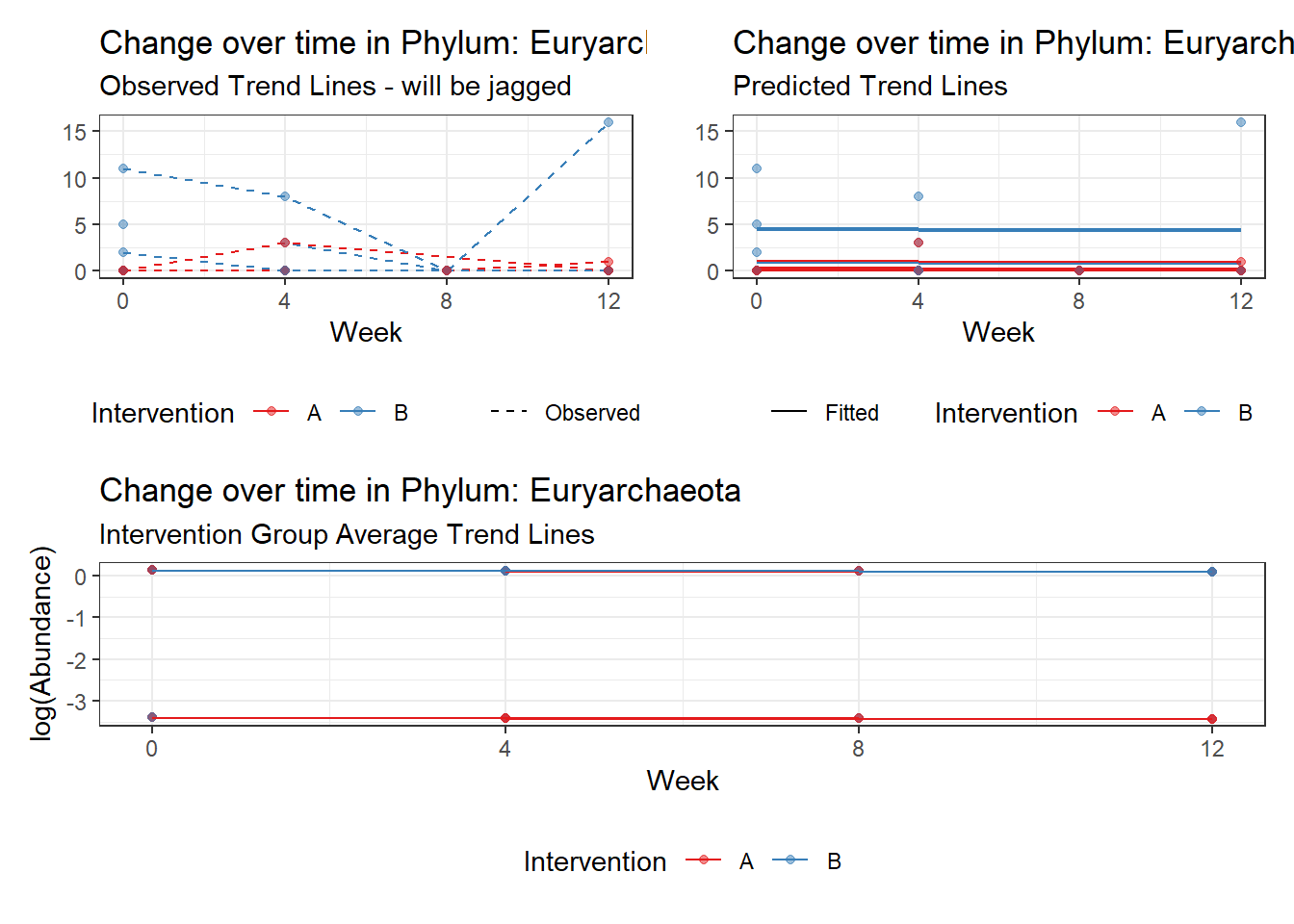
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -3.39308 1.36474 -2.48624 0.01291
time Euryarchaeota -0.01230 0.13243 -0.09285 0.92602
hispanic Euryarchaeota 3.53265 1.41244 2.50110 0.01238
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.3207
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.1316301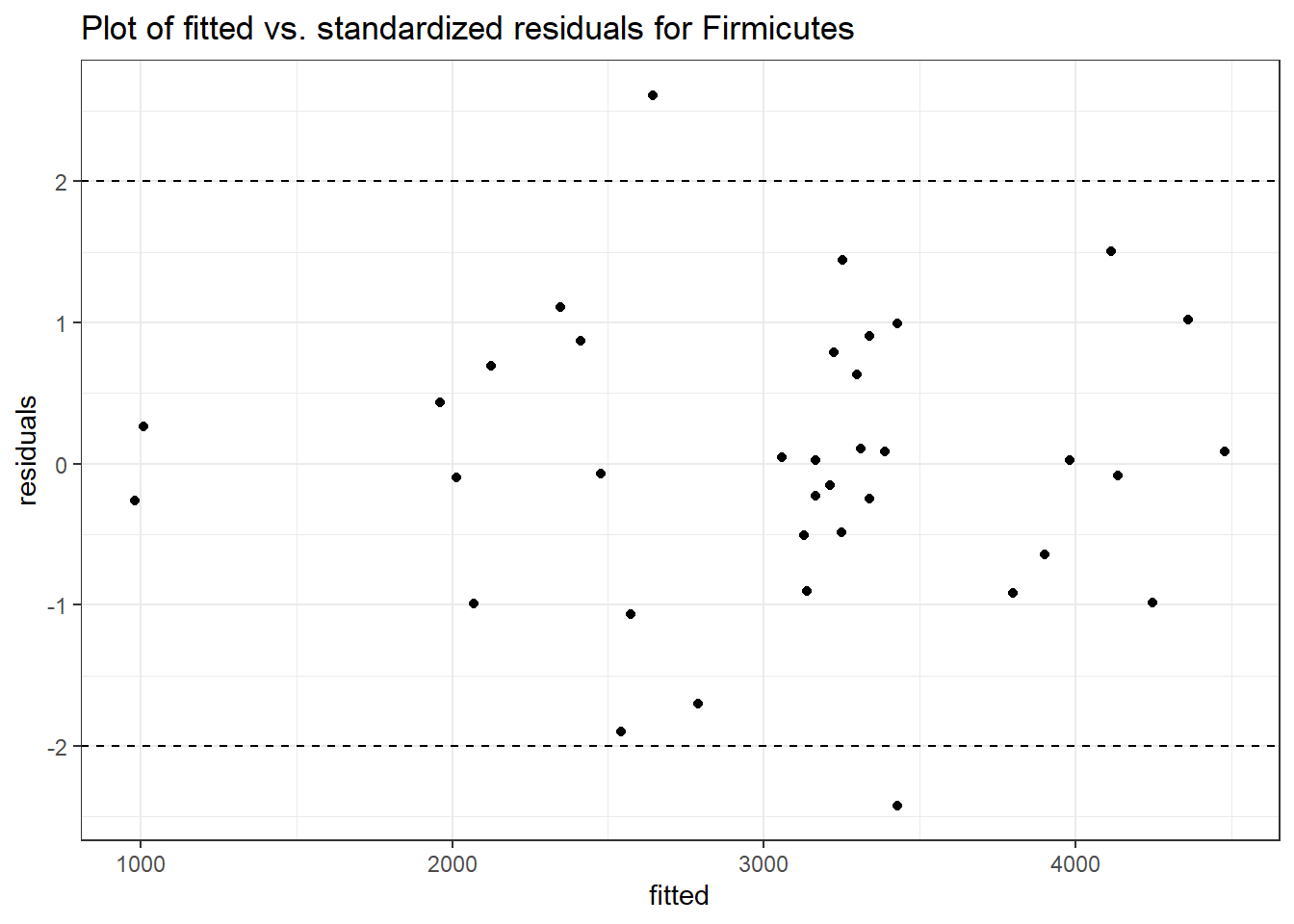
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.87986 0.19477 40.45704 0.00000
time Firmicutes 0.02657 0.00267 9.96905 0.00000
hispanic Firmicutes 0.06564 0.24413 0.26889 0.78801
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.38934
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.8242502

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae -1.36816 1.32622 -1.03162 0.30225
time Lentisphaerae 0.30516 0.05828 5.23622 0.00000
hispanic Lentisphaerae 1.99760 1.52540 1.30956 0.19035
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1656
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.06821181

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.94109 0.13653 43.51361 0.00000
time Proteobacteria -0.04985 0.00838 -5.95264 0.00000
hispanic Proteobacteria -0.28727 0.17084 -1.68152 0.09266
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.27056
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.8933541
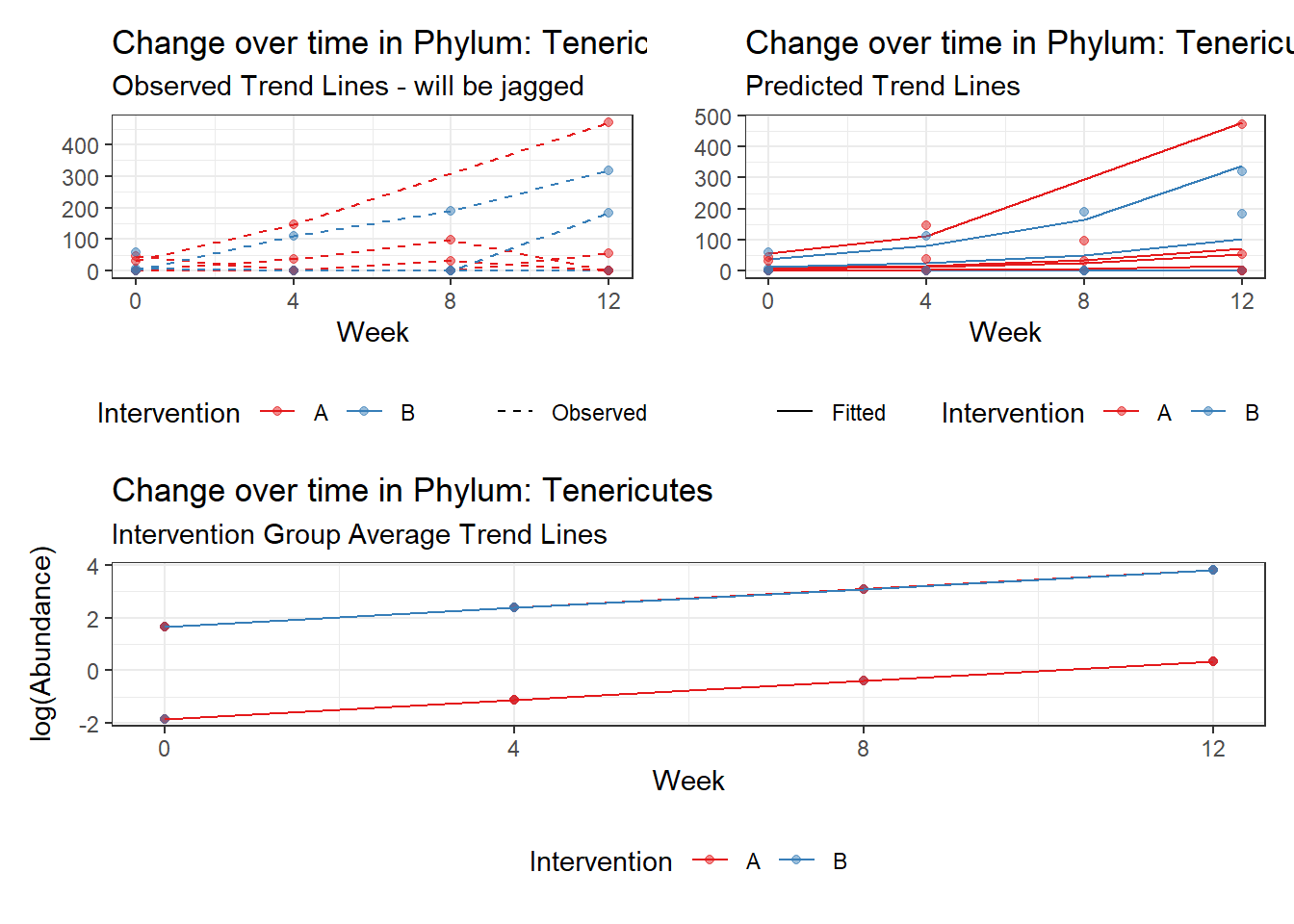
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes -1.84391 1.71374 -1.07596 0.28195
time Tenericutes 0.72216 0.02492 28.98177 0.00000
hispanic Tenericutes 3.49256 2.01726 1.73134 0.08339
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.8943
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000004ce5a200>
Link: poisson
Intraclass Correlation (ICC): 0.8512603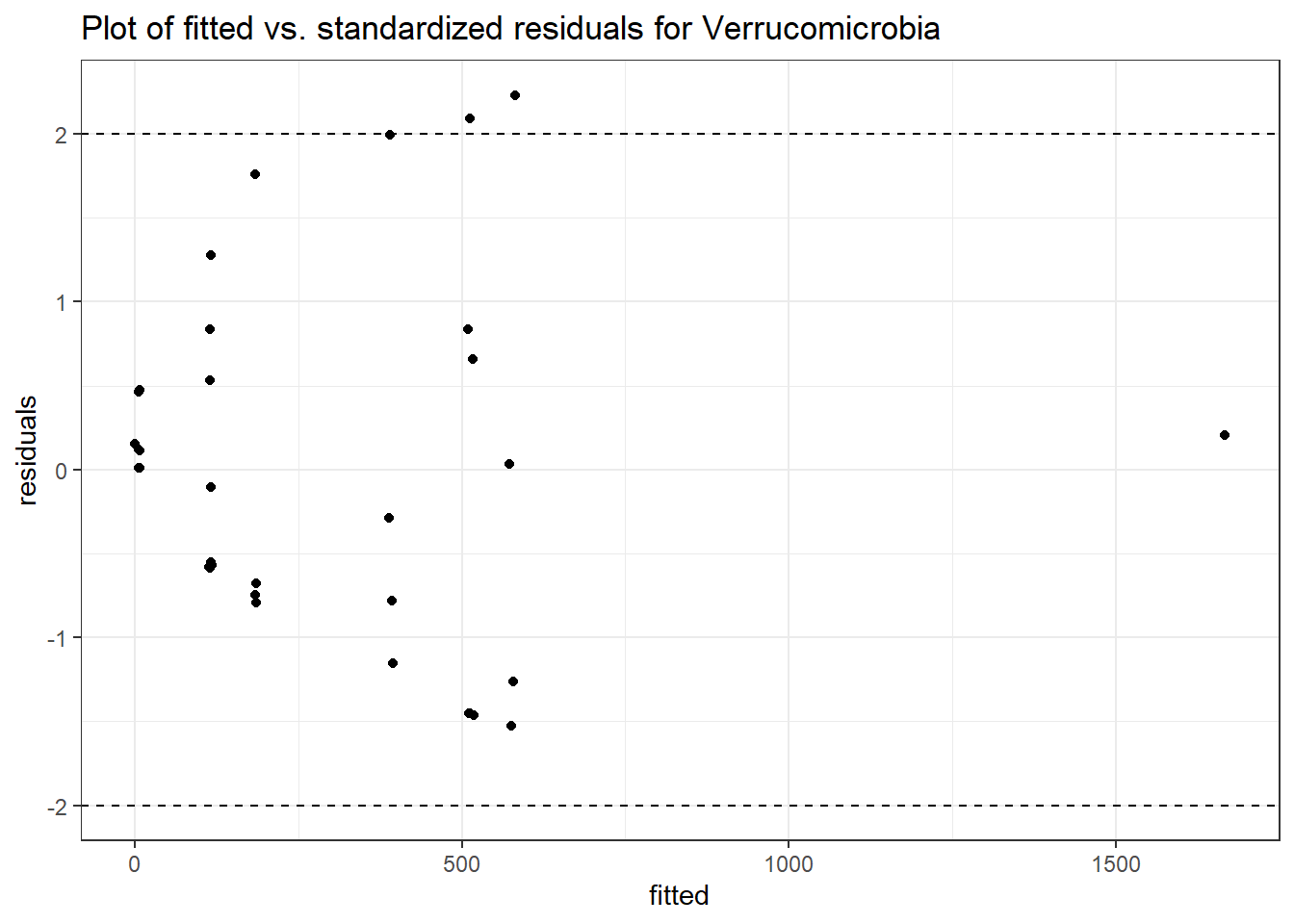

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 3.81822 1.19875 3.18516 0.00145
time Verrucomicrobia 0.00501 0.00920 0.54458 0.58604
hispanic Verrucomicrobia 1.09275 1.50857 0.72436 0.46884
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3923
Significance test not available for random effectsphylum.fit3b <- glmm_microbiome(mydata=microbiome_data,model.number=3,
taxa.level="Phylum", link="negbinom",
model="1 + time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000003ebecac0>
Link: negbinom
Intraclass Correlation (ICC): 0.3725376
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.16720 0.58413 3.71010 0.00021
time Actinobacteria 0.22287 0.19061 1.16922 0.24232
hispanic Actinobacteria 0.48407 0.63776 0.75902 0.44784
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.77053
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000003ebecac0>
Link: negbinom
Intraclass Correlation (ICC): 0.1236759
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.25335 0.20482 40.29657 0.00000
time Bacteroidetes -0.04826 0.03488 -1.38335 0.16656
hispanic Bacteroidetes -0.32376 0.25090 -1.29040 0.19691
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.37567
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000003ebecac0>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0157651 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 9.048682e-13
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -2.41570 2.55871 -0.94411 0.34511
time Cyanobacteria 1.31650 0.89582 1.46959 0.14167
hispanic Cyanobacteria 4.89246 2.19467 2.22924 0.02580
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 9.5125e-07
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000003ebecac0>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 2.303365e-10
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -3.04341 1.47908 -2.05764 0.03962
time Euryarchaeota 0.16964 0.42605 0.39816 0.69051
hispanic Euryarchaeota 3.62365 1.38726 2.61210 0.00900
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.5177e-05
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000003ebecac0>
Link: negbinom
Intraclass Correlation (ICC): 0.1198663
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.88771 0.19223 41.03337 0.00000
time Firmicutes 0.02203 0.02412 0.91332 0.36107
hispanic Firmicutes 0.06340 0.23817 0.26619 0.79010
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.36904
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000003ebecac0>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 2.414581e-10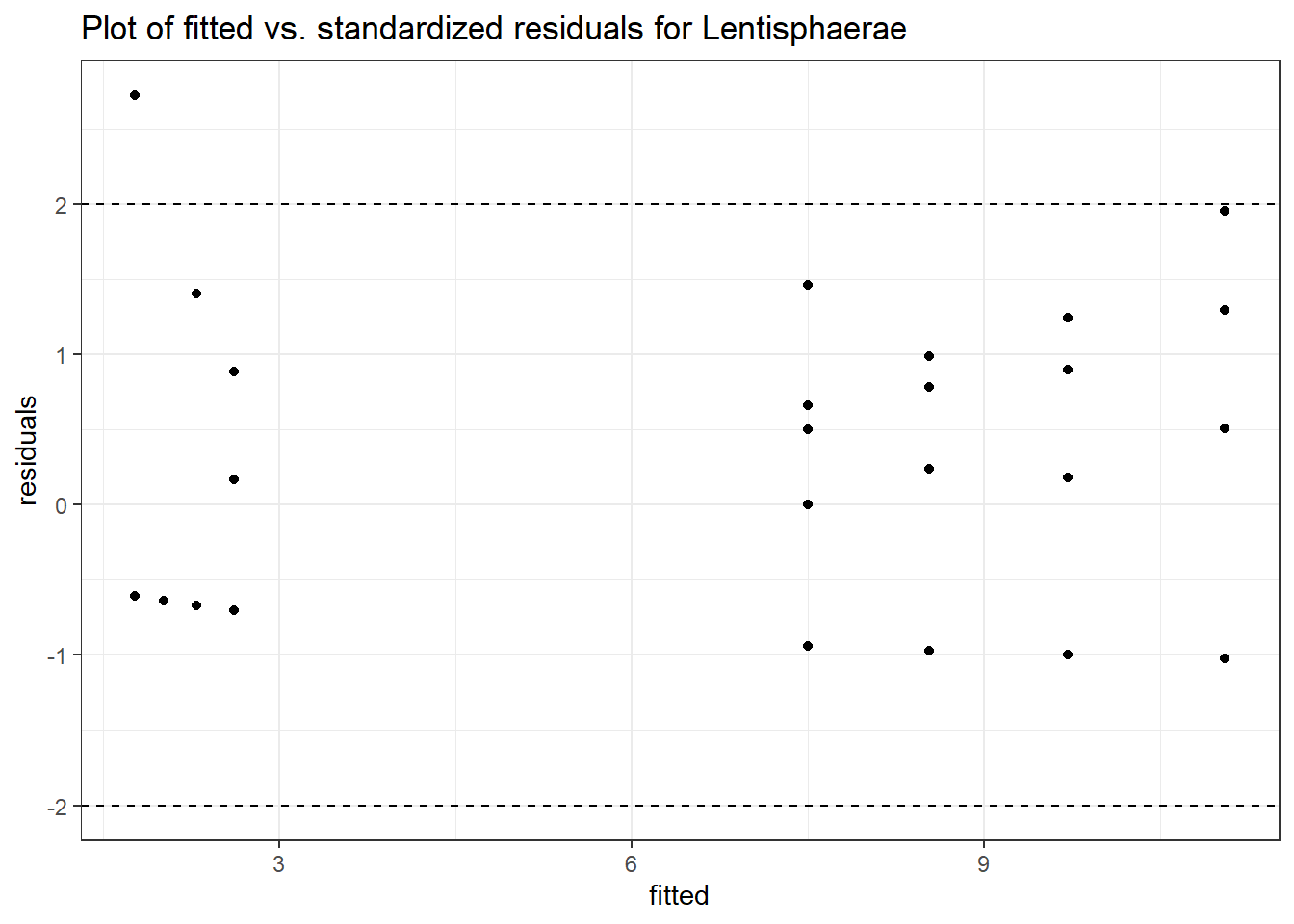
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.57149 0.63203 0.90422 0.36588
time Lentisphaerae 0.12932 0.29903 0.43247 0.66540
hispanic Lentisphaerae 1.44284 0.78080 1.84790 0.06462
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.5539e-05
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000003ebecac0>
Link: negbinom
Intraclass Correlation (ICC): 0.008002456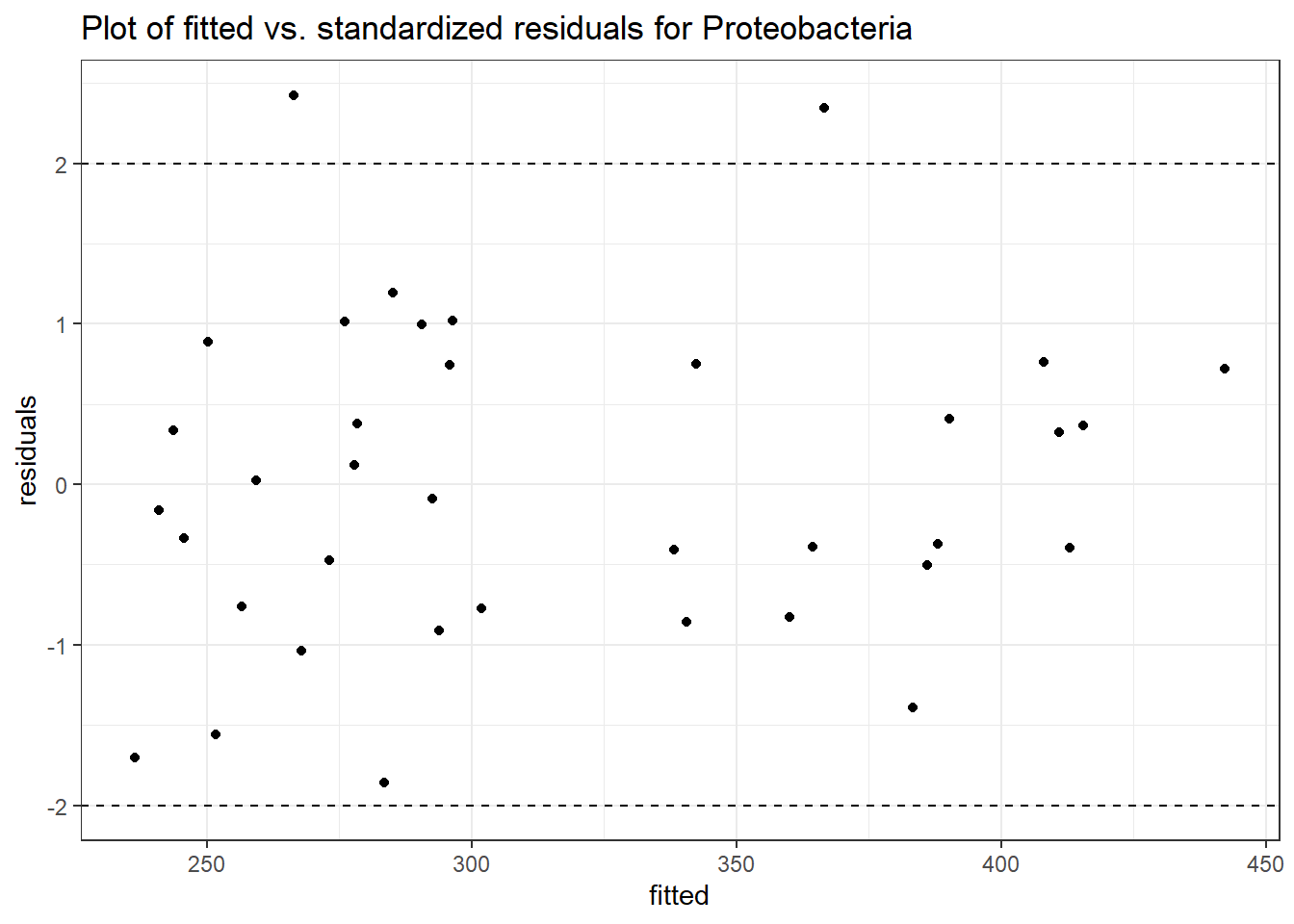
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.03616 0.21207 28.46316 0.00000
time Proteobacteria -0.06249 0.08890 -0.70300 0.48205
hispanic Proteobacteria -0.35349 0.21780 -1.62302 0.10459
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.089817
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x000000003ebecac0>
Link: negbinomError in factory(refitNB, types = c("message", "warning"))(lastfit, theta = exp(t), : pwrssUpdate did not converge in (maxit) iterations
Controlling for the effect of time, Ethnicity was not significant for all Phylum.
Effect of Intervention
Next, we invested the effect of intervention, however, the results are parsed between which covariates are included in the model.
Intervention was coded as (group A = 0, and group B = 1). So, the results are interpretted as the effect of being in intervention group B.
Covariate(s): Time
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Intervention)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
The effect of intervention is the main outcome of this study.
phylum.fit4 <- glmm_microbiome(mydata=microbiome_data,model.number=4,
taxa.level="Phylum", link="poisson",
model="1 + intB + time + (1|SubjectID)")
#########################################
#########################################
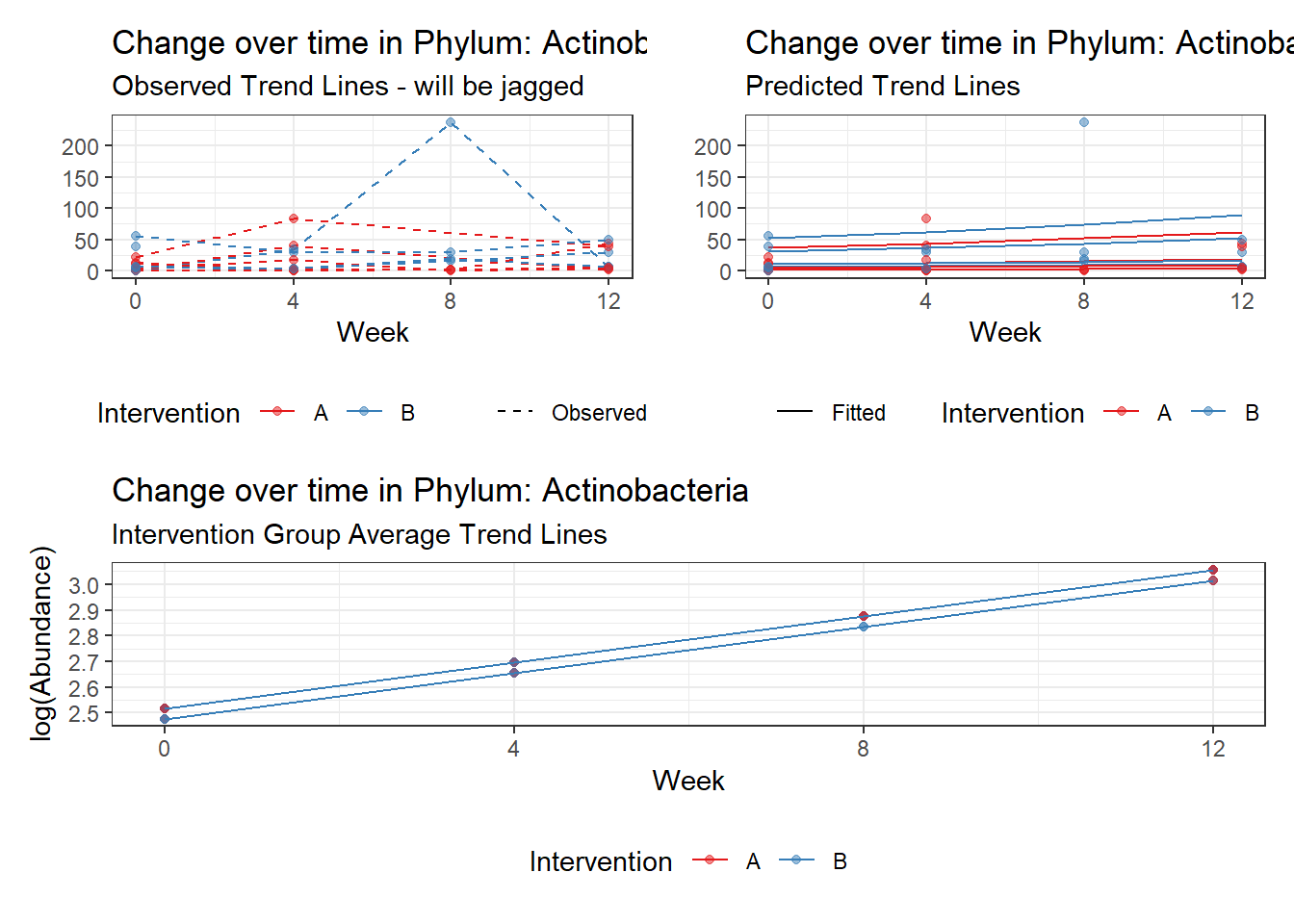
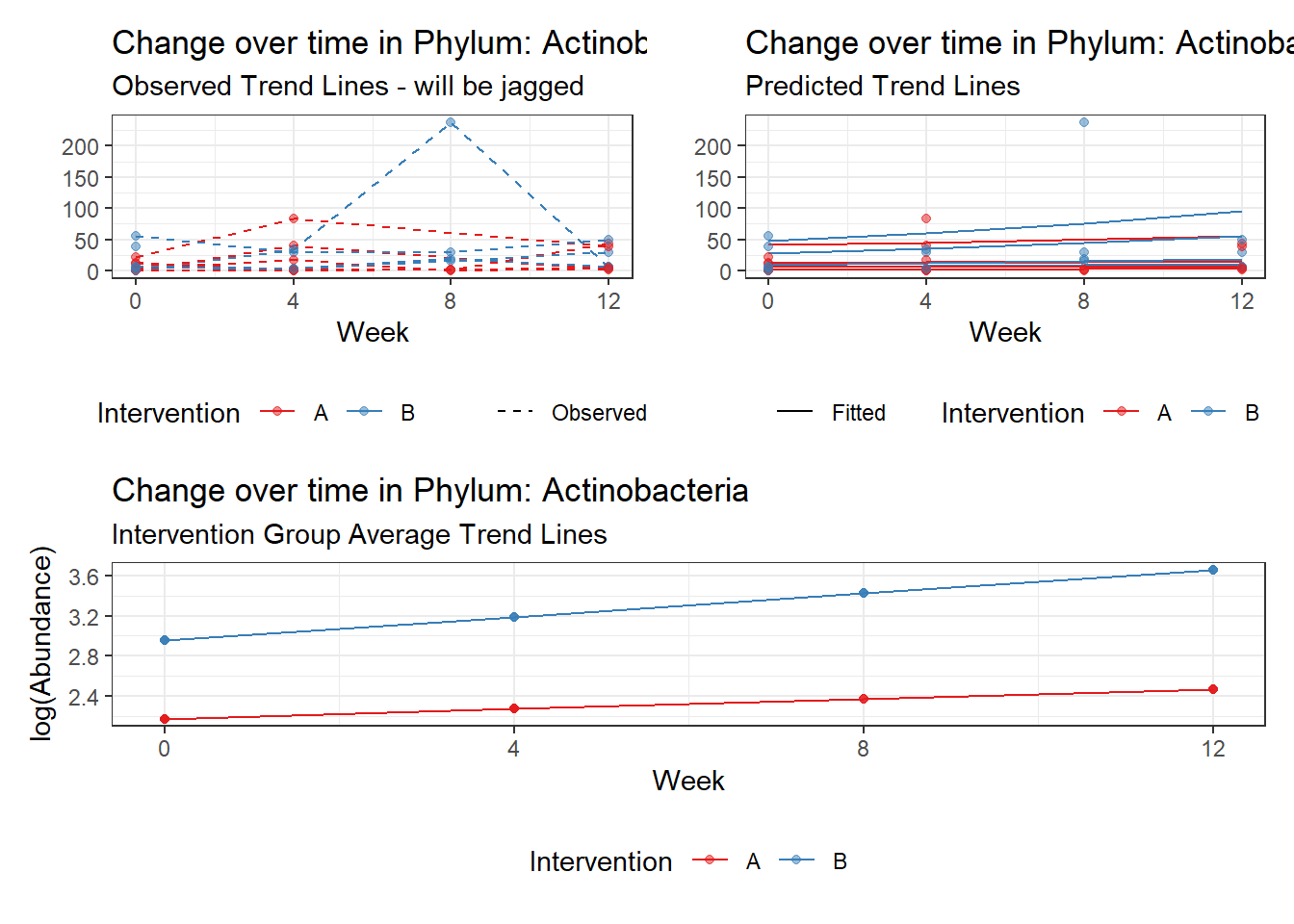
Model: Actinobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.4323474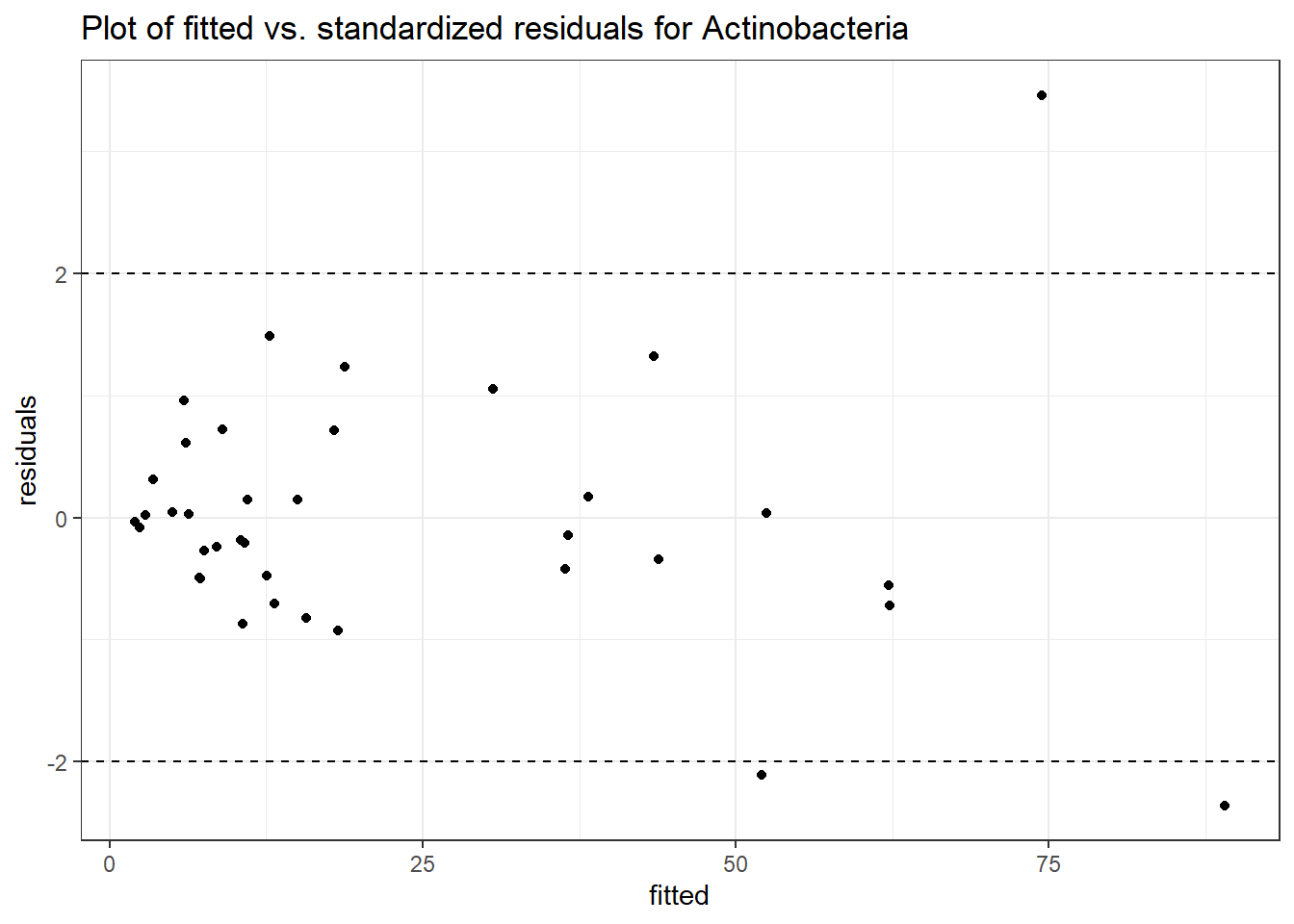
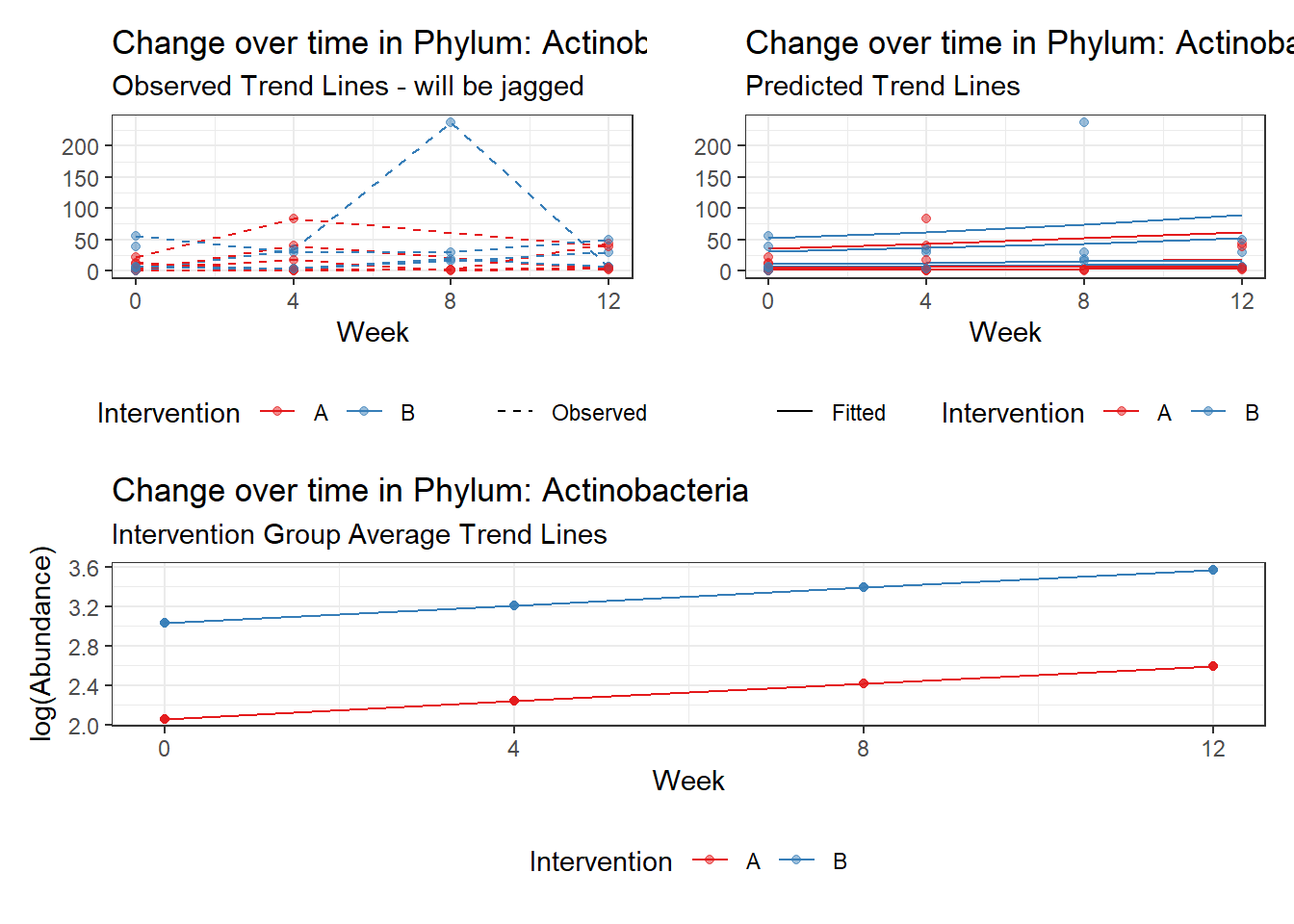
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.05964 0.36887 5.58362 0.00000
intB Actinobacteria 0.97330 0.53808 1.80883 0.07048
time Actinobacteria 0.17910 0.03052 5.86852 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.87272
Significance test not available for random effects
#########################################
#########################################
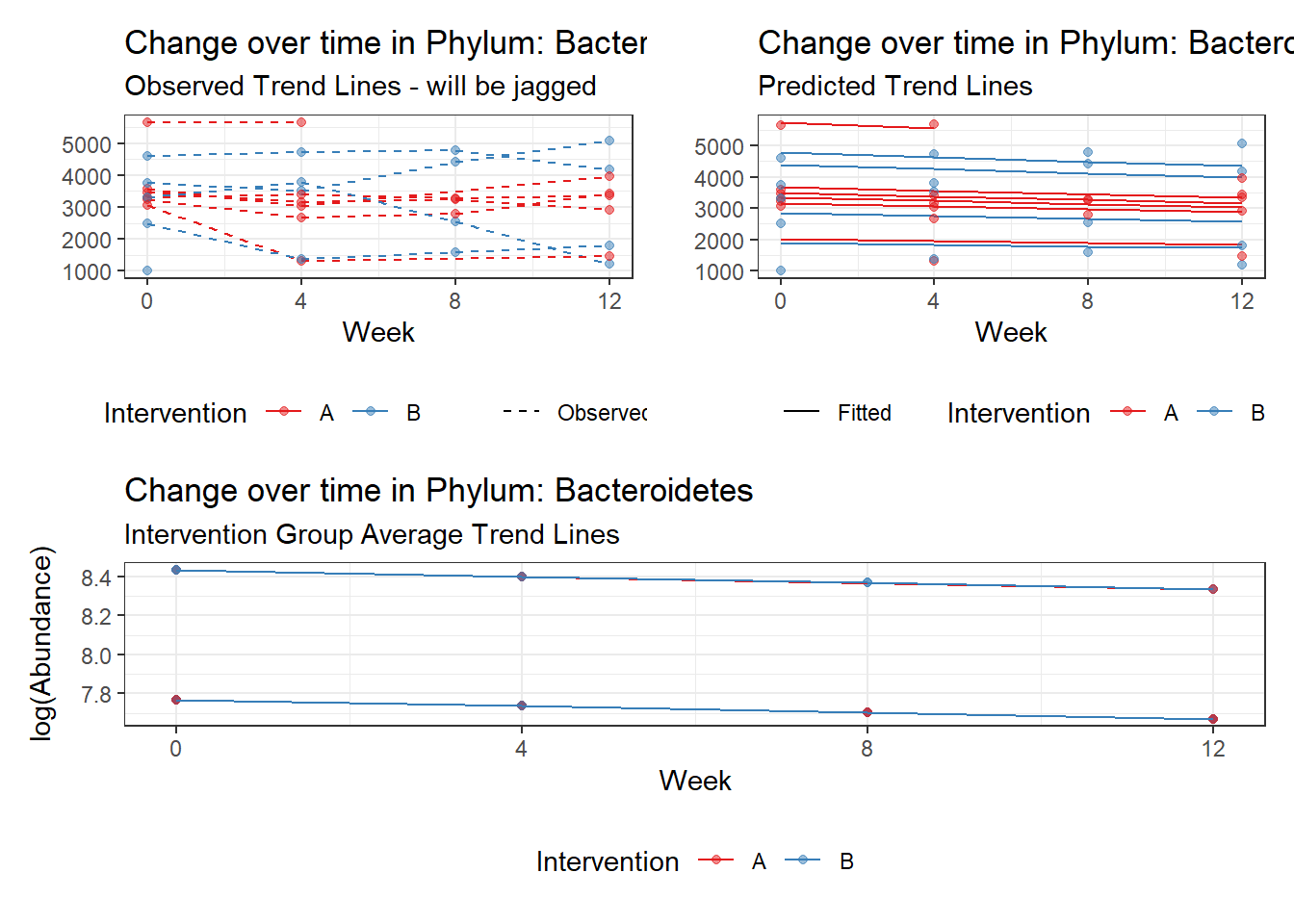
Model: Bacteroidetes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.1676984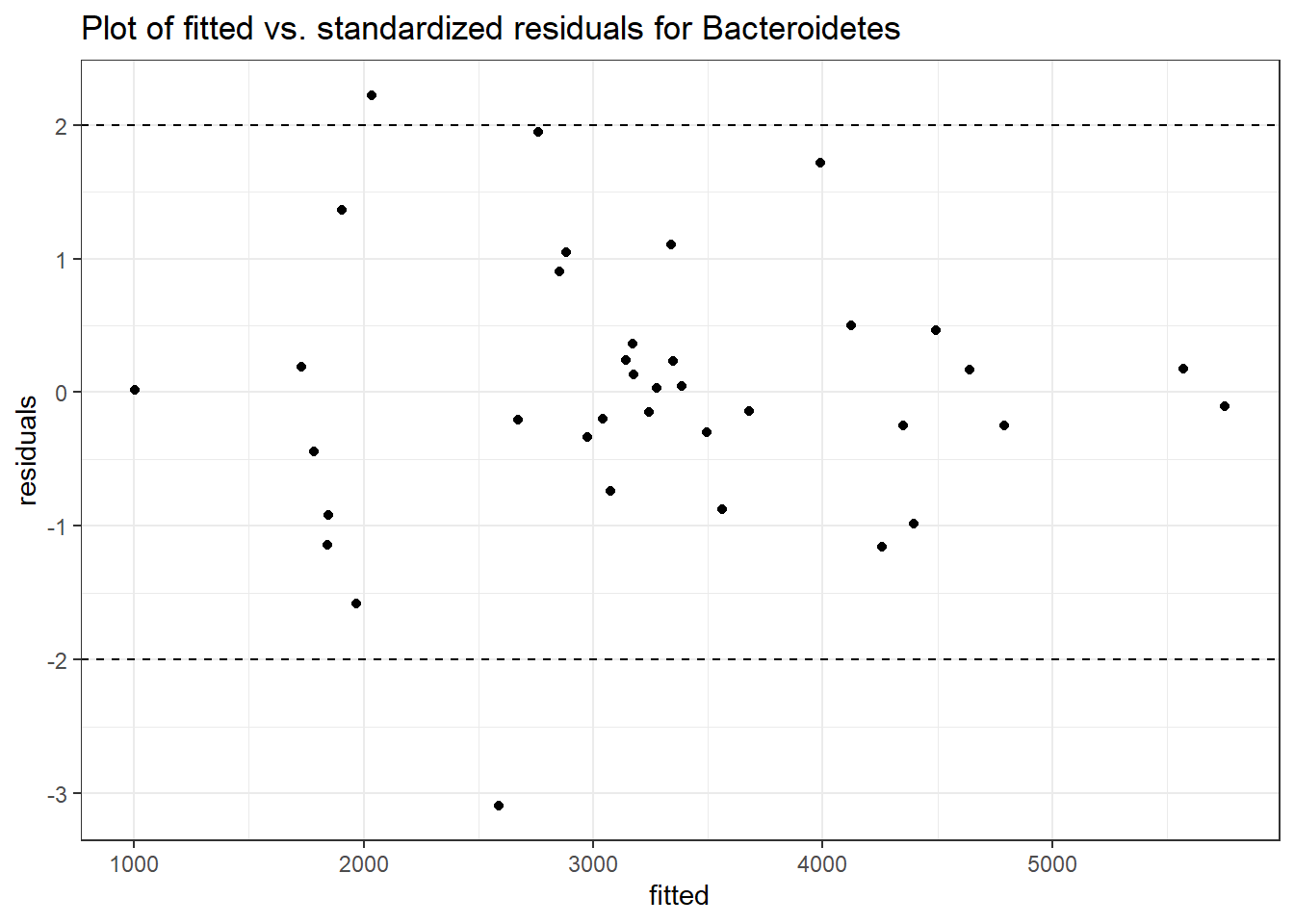
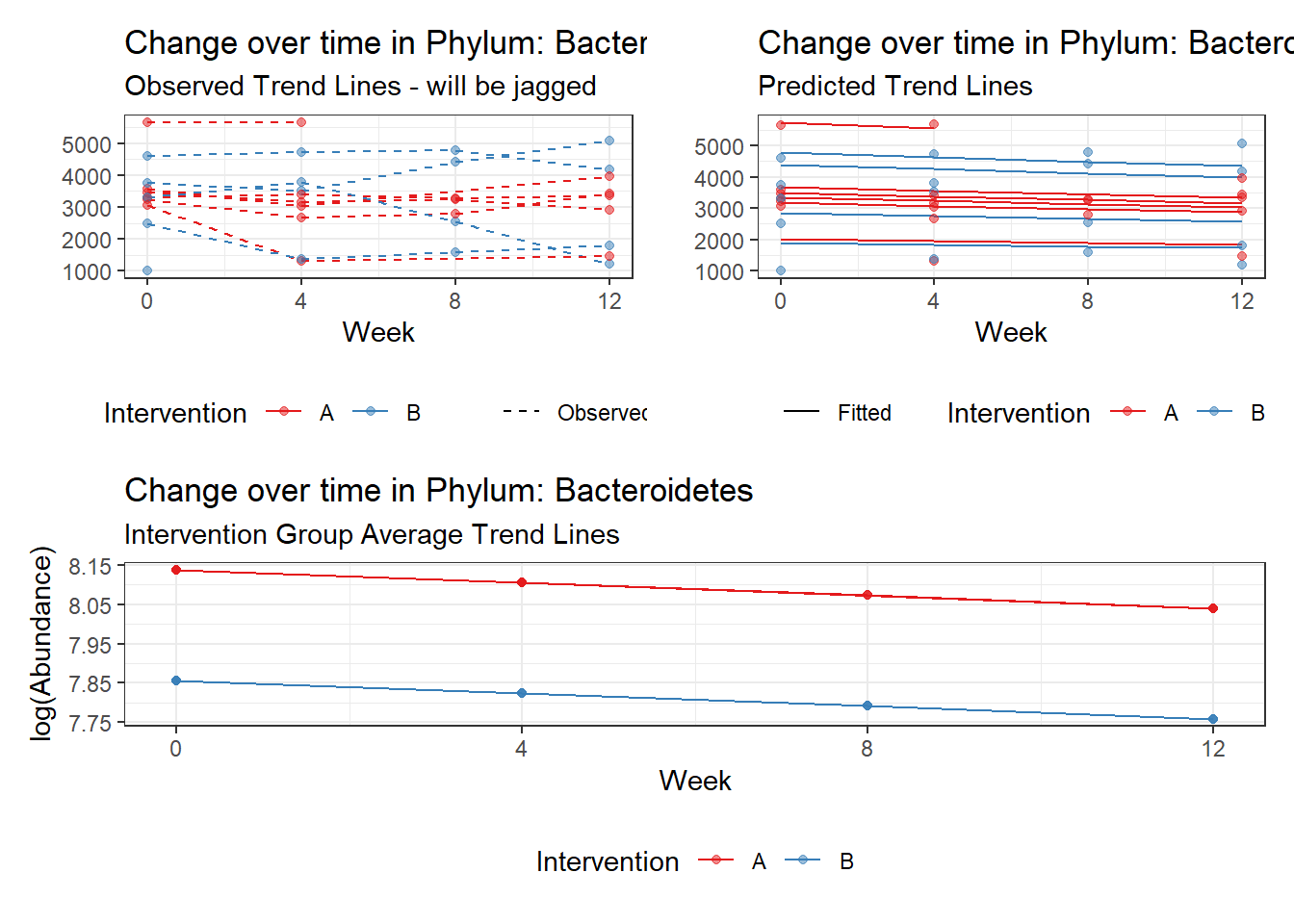
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.13729 0.18333 44.38648 0.00000
intB Bacteroidetes -0.28105 0.27194 -1.03351 0.30137
time Bacteroidetes -0.03231 0.00265 -12.17884 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.44887
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.9441699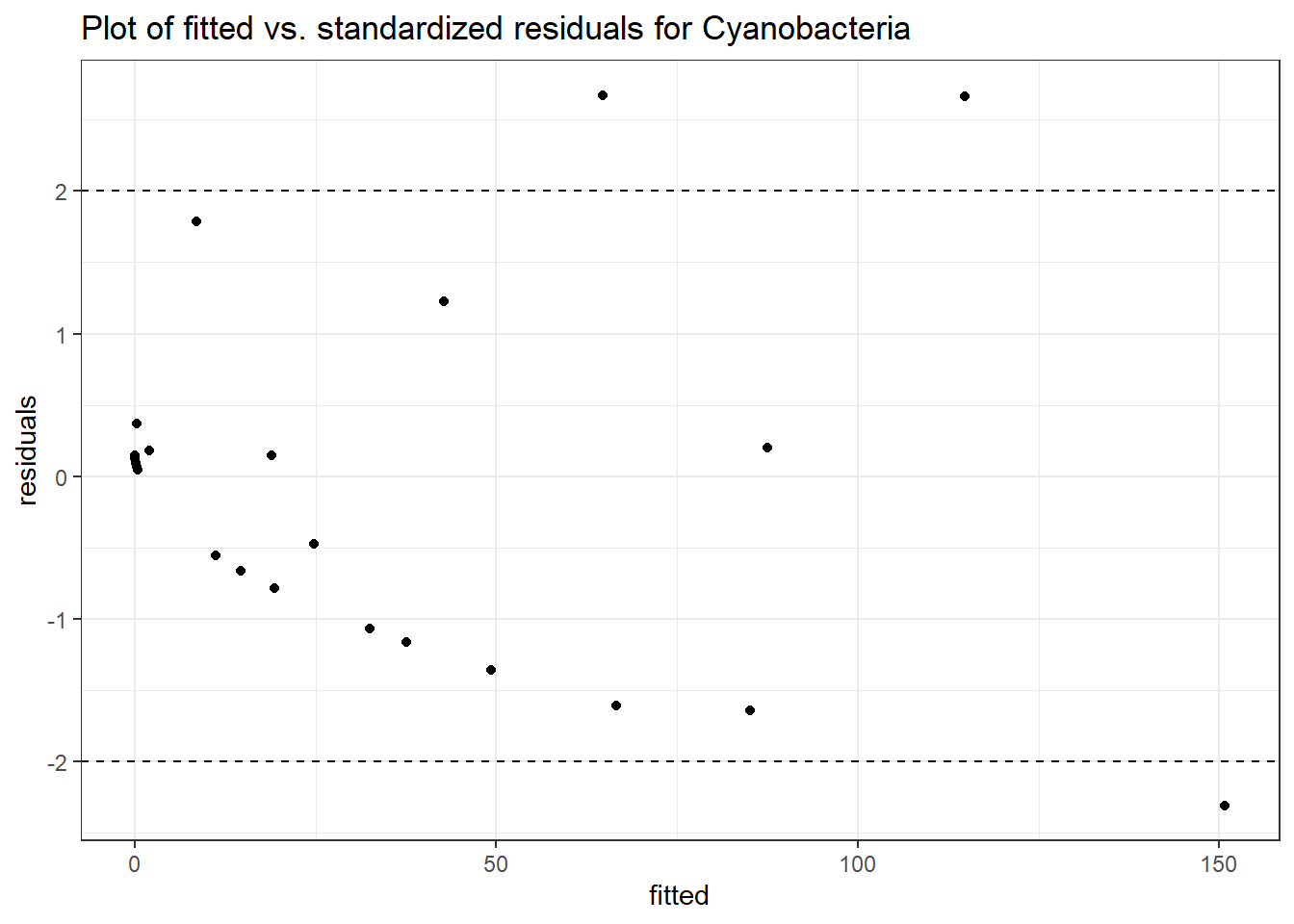
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -1.43605 2.23406 -0.64280 0.52035
intB Cyanobacteria 1.37007 2.85794 0.47939 0.63166
time Cyanobacteria -0.27252 0.03201 -8.51438 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.1124
Significance test not available for random effects
#########################################
#########################################
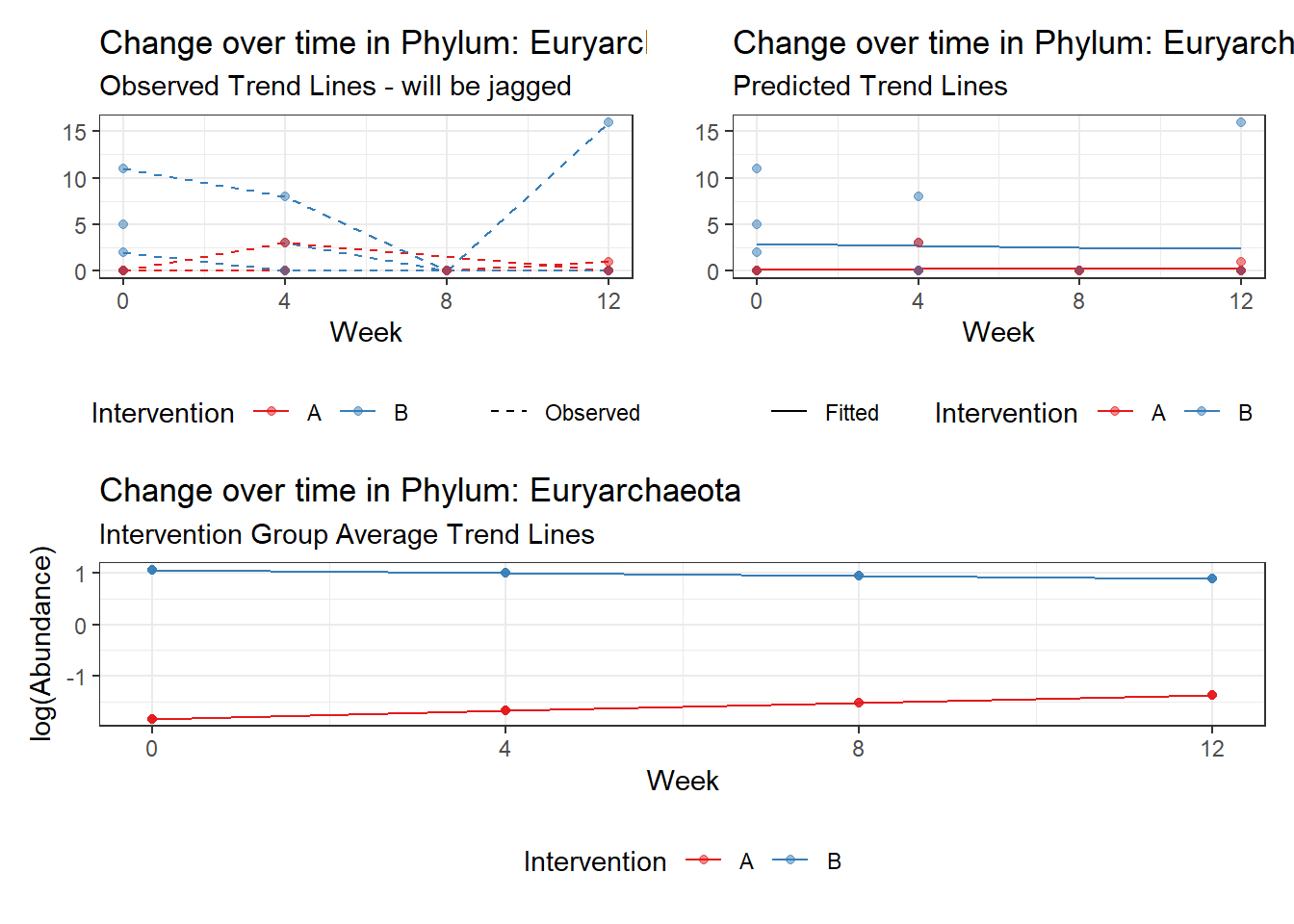
Model: Euryarchaeota ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.6298254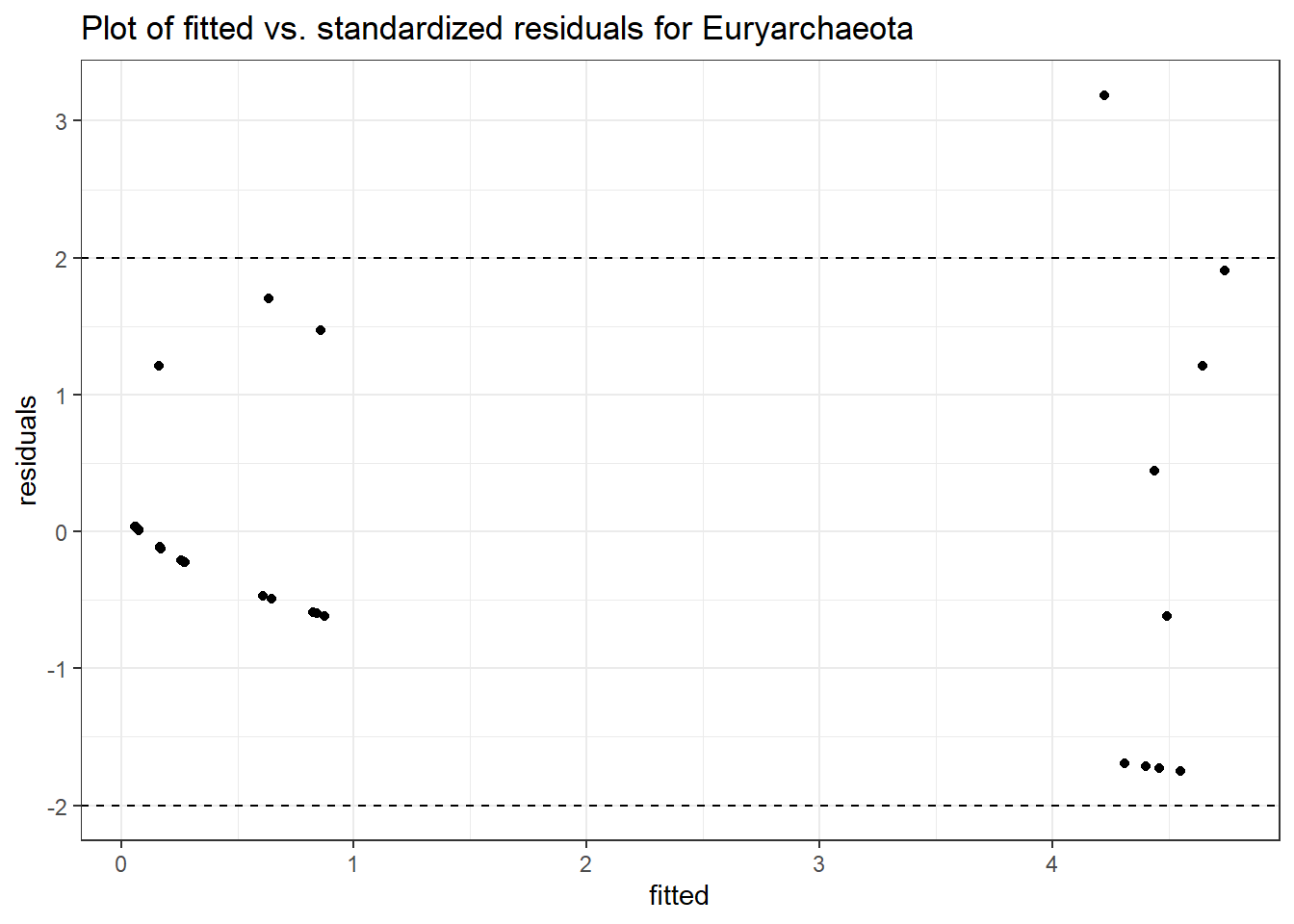
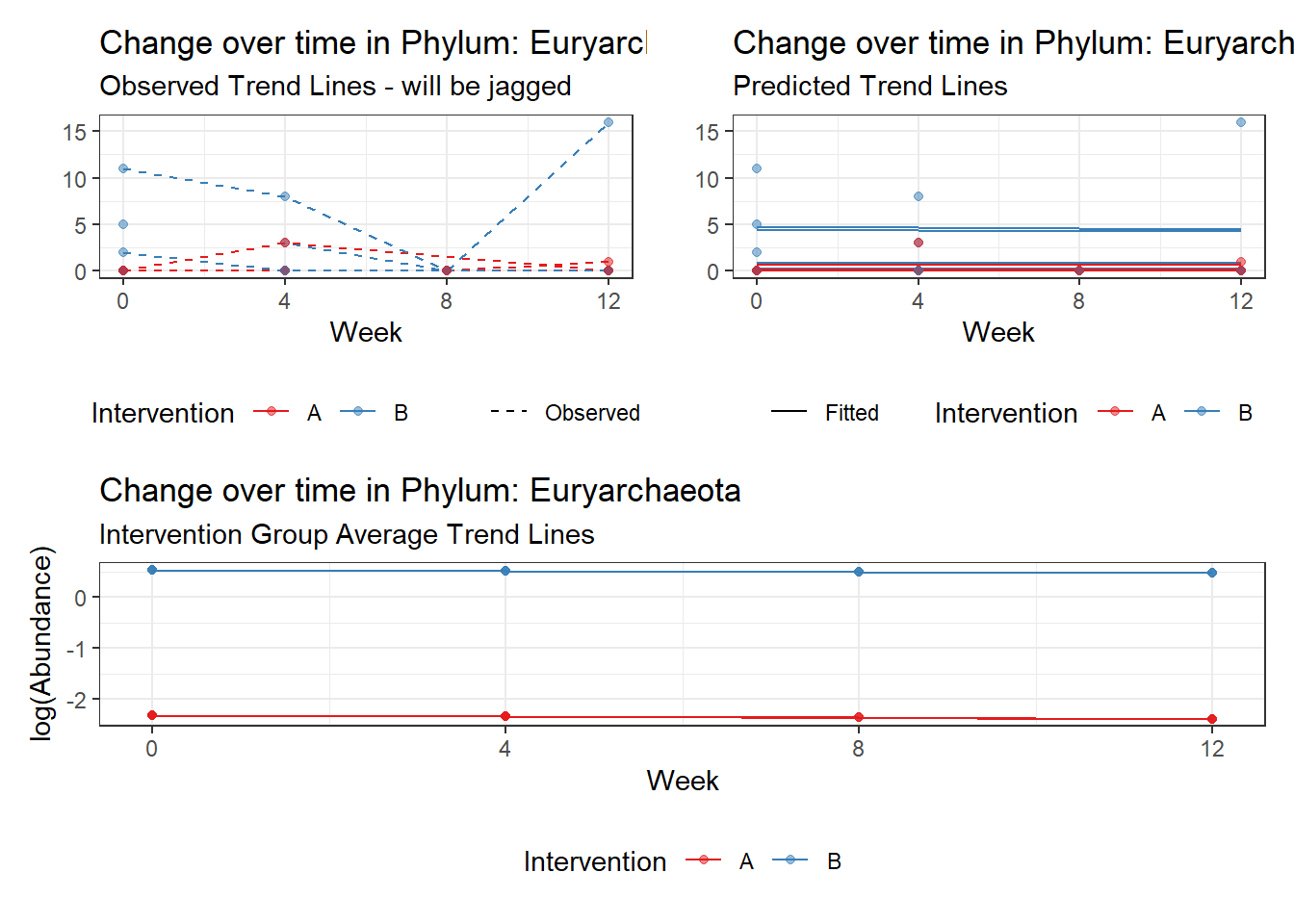
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -2.32923 0.96087 -2.42408 0.01535
intB Euryarchaeota 2.86692 1.07899 2.65704 0.00788
time Euryarchaeota -0.02057 0.13259 -0.15513 0.87672
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.3044
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.1283903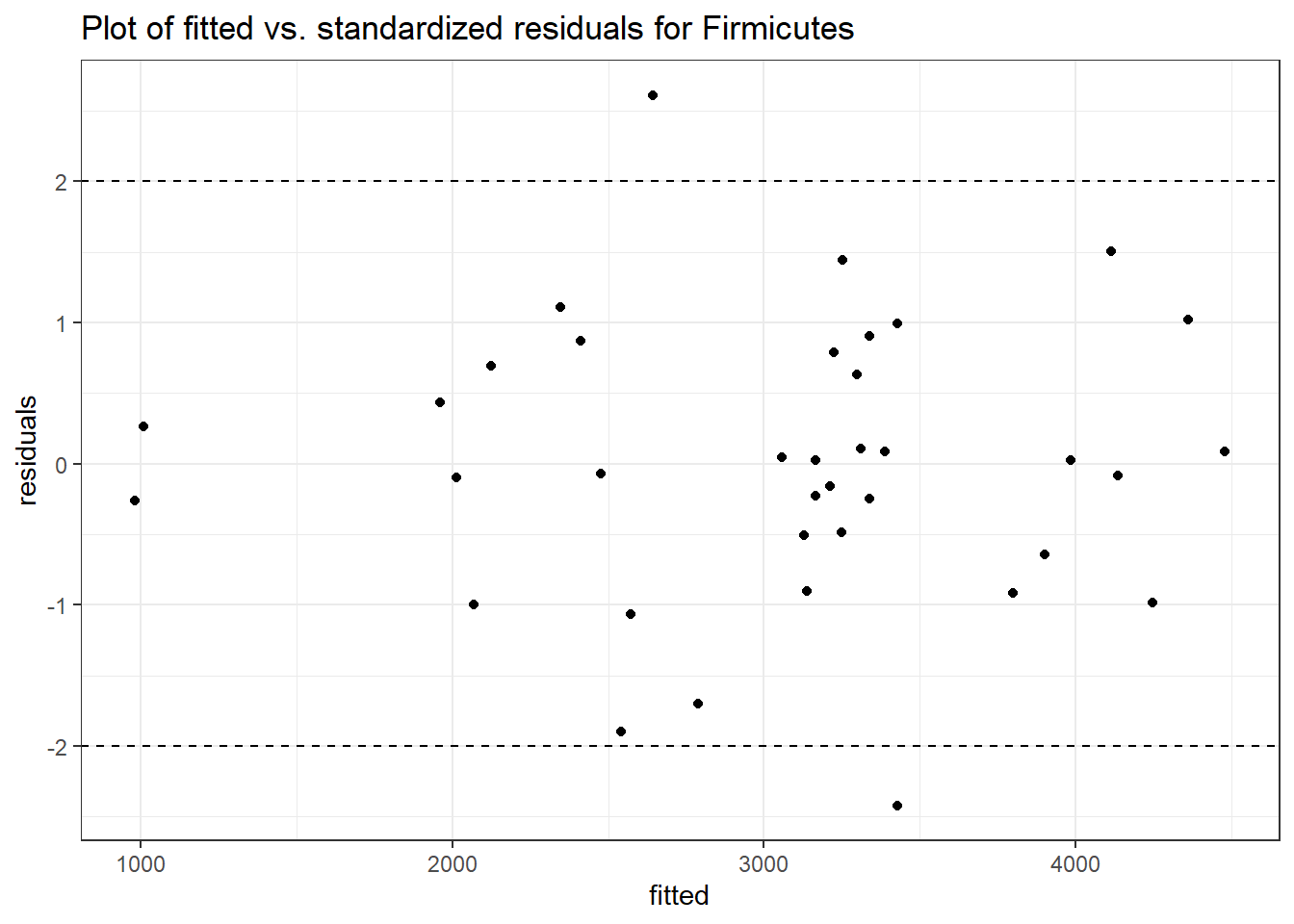
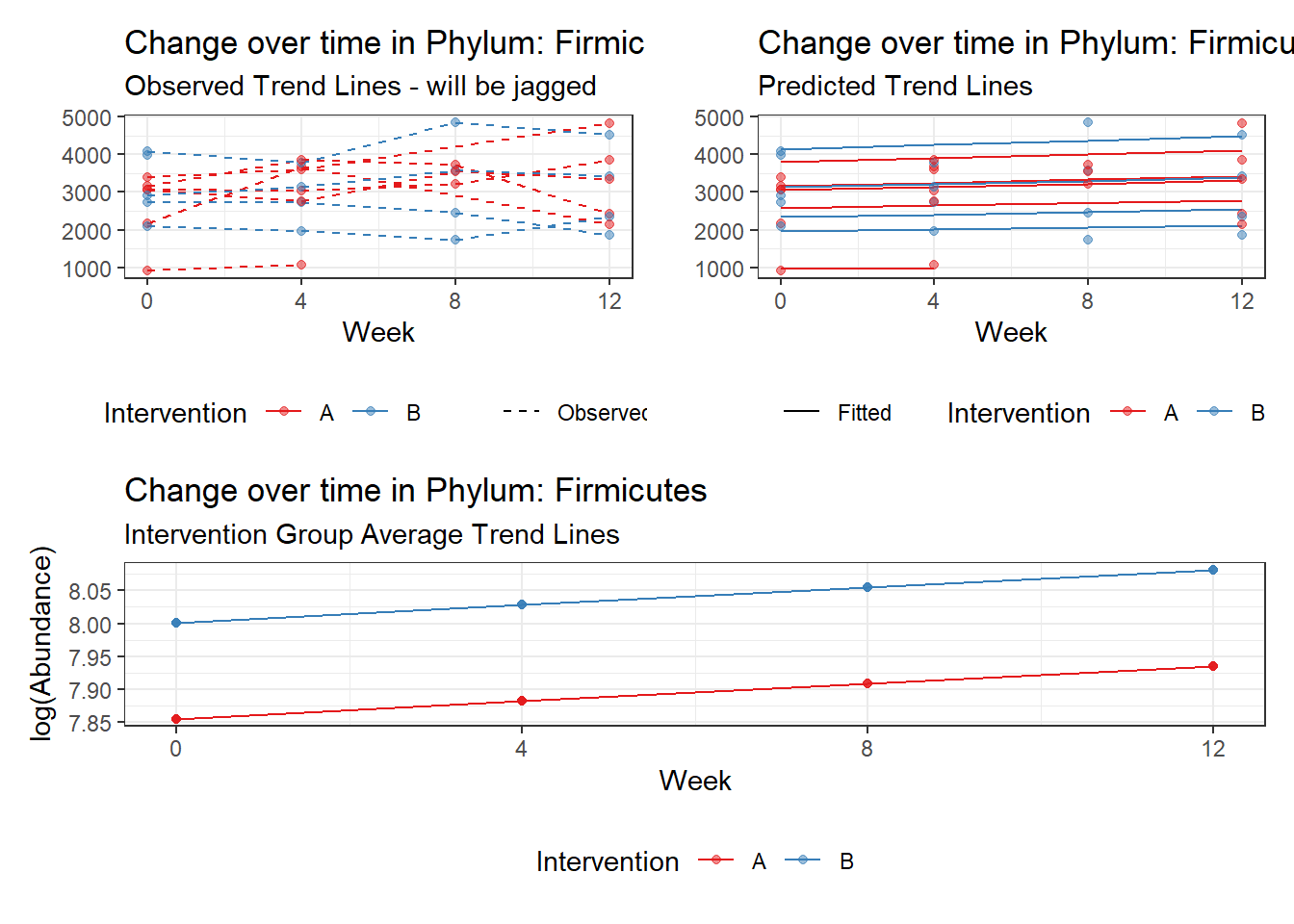
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.85508 0.15681 50.09361 0.00000
intB Firmicutes 0.14637 0.23251 0.62952 0.52901
time Firmicutes 0.02657 0.00267 9.96894 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.3838
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.8556082
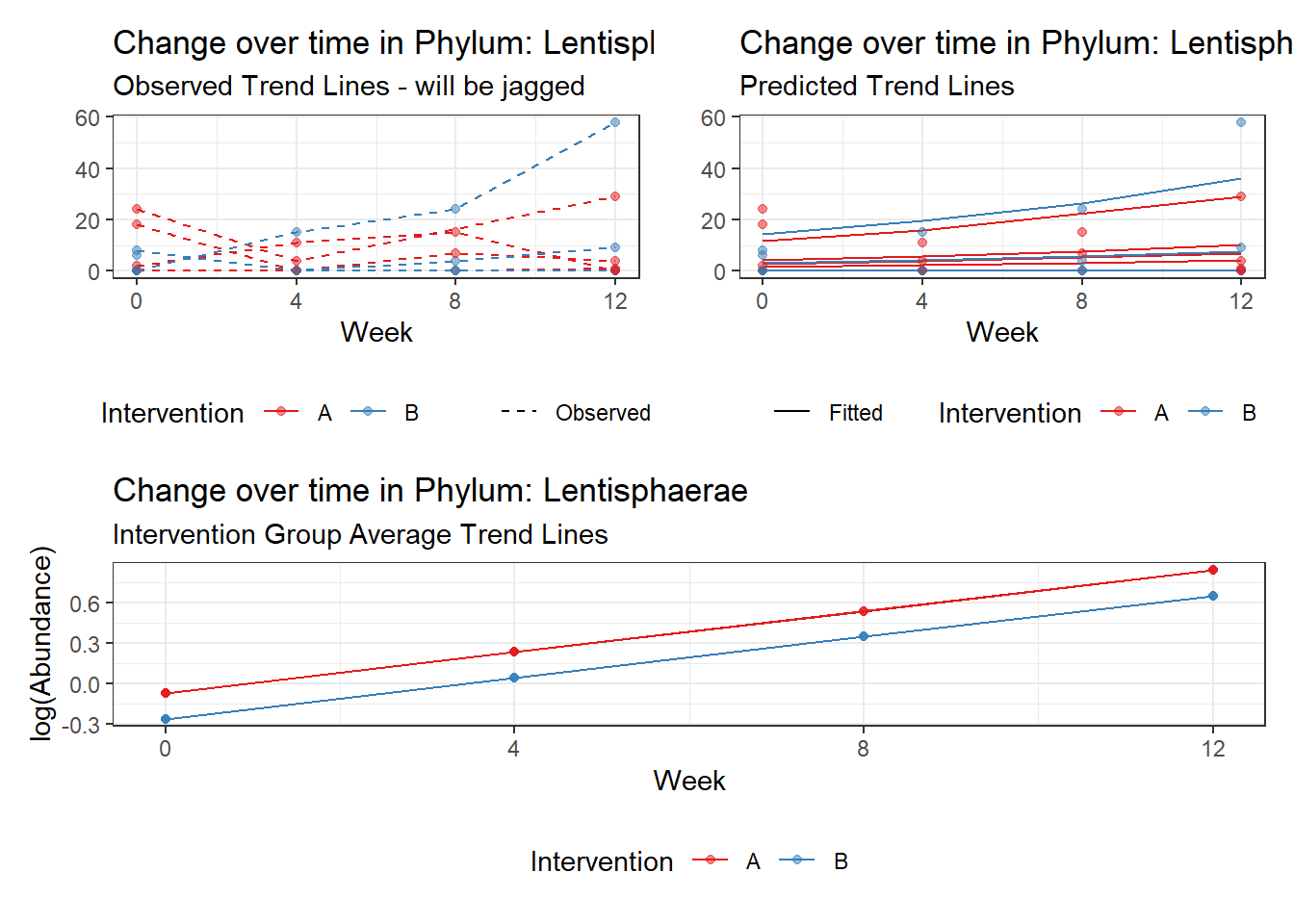
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae -0.07292 1.11828 -0.06520 0.94801
intB Lentisphaerae -0.18831 1.59508 -0.11805 0.90602
time Lentisphaerae 0.30388 0.05824 5.21735 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4343
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.07459268
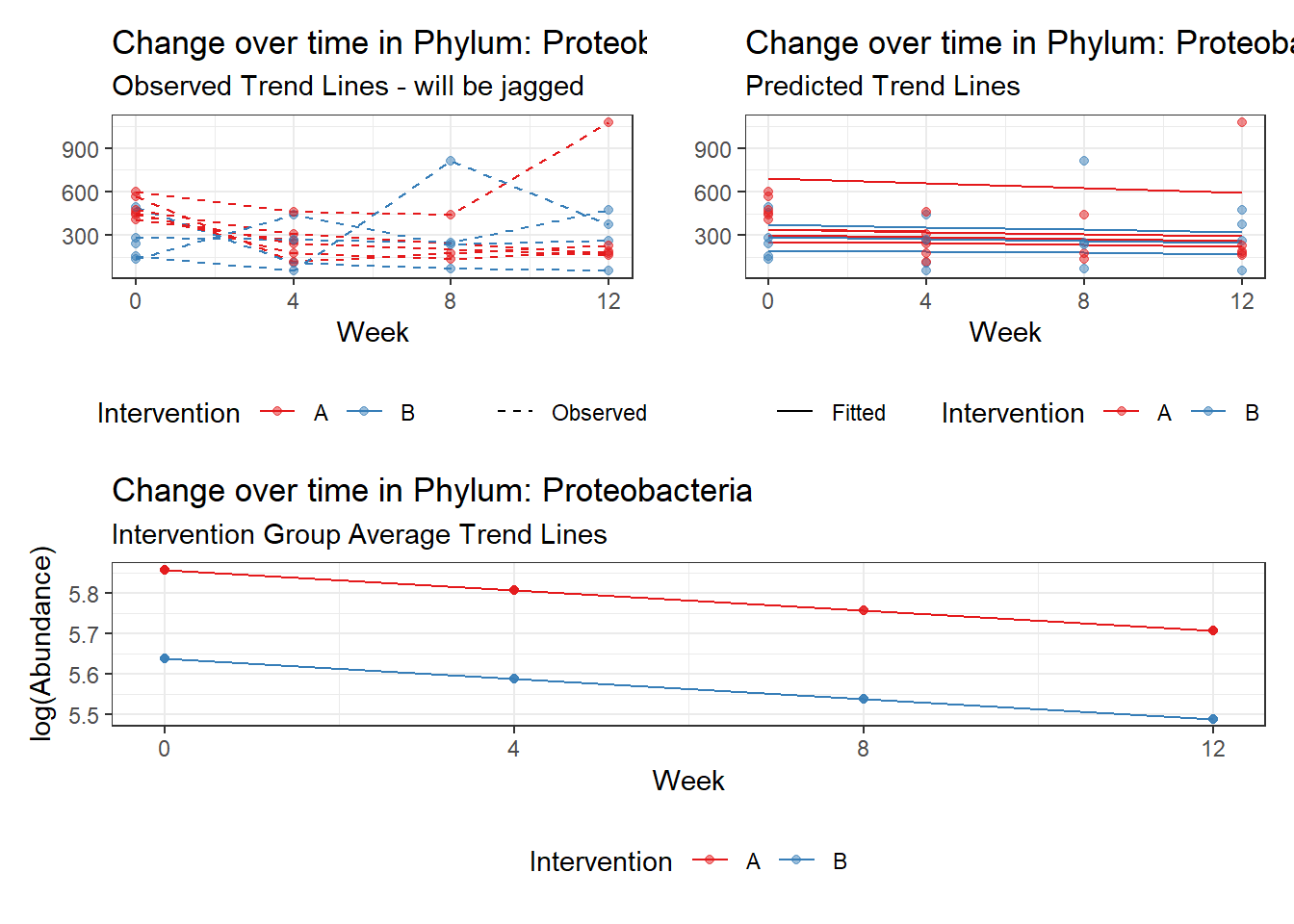
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.85740 0.11708 50.03022 0.00000
intB Proteobacteria -0.21827 0.17331 -1.25945 0.20787
time Proteobacteria -0.04969 0.00838 -5.93286 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.28391
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.9160166
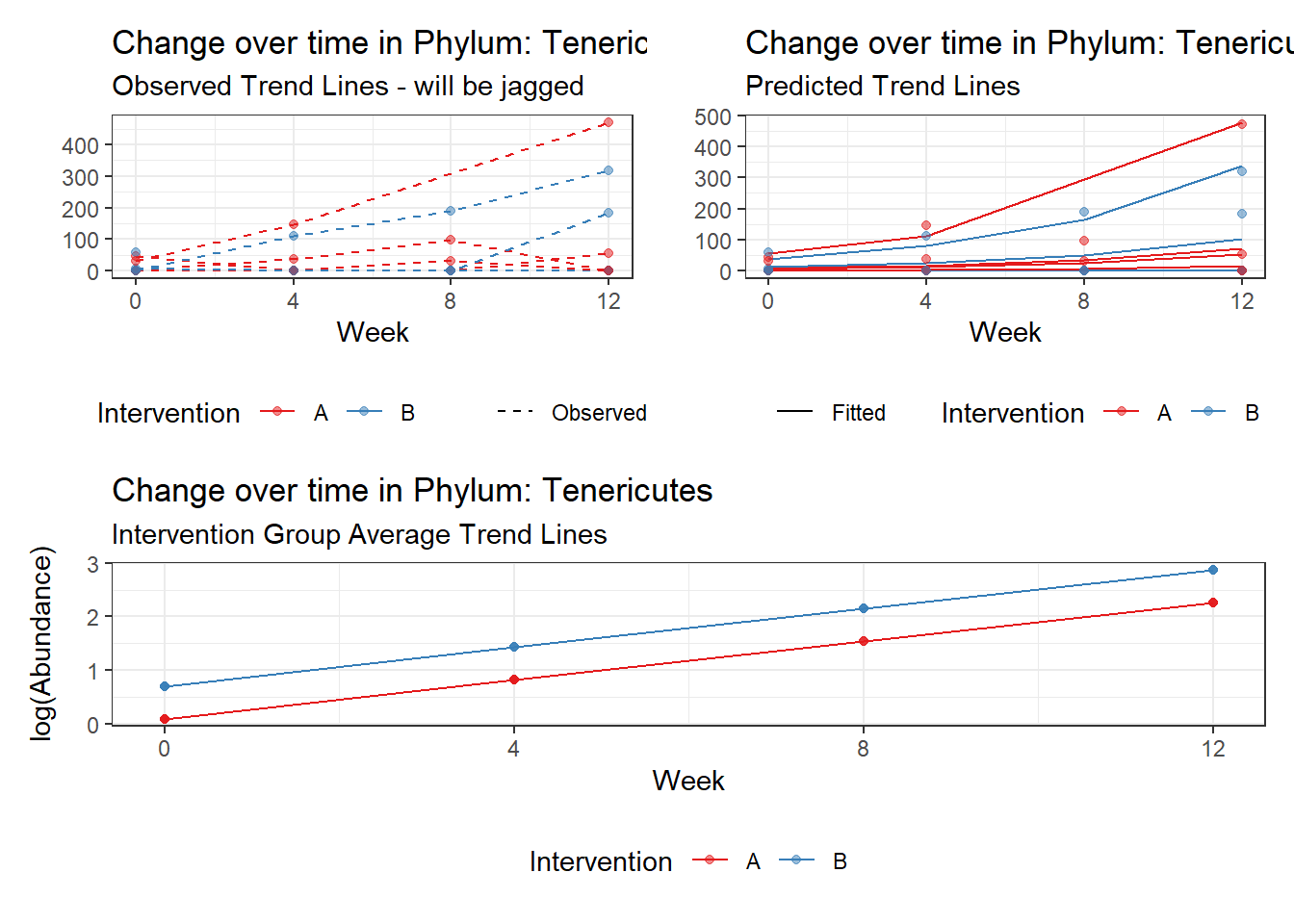
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 0.09290 1.40717 0.06602 0.94736
intB Tenericutes 0.60940 2.05410 0.29668 0.76671
time Tenericutes 0.72195 0.02489 29.01032 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.3026
Significance test not available for random effects
#########################################
#########################################
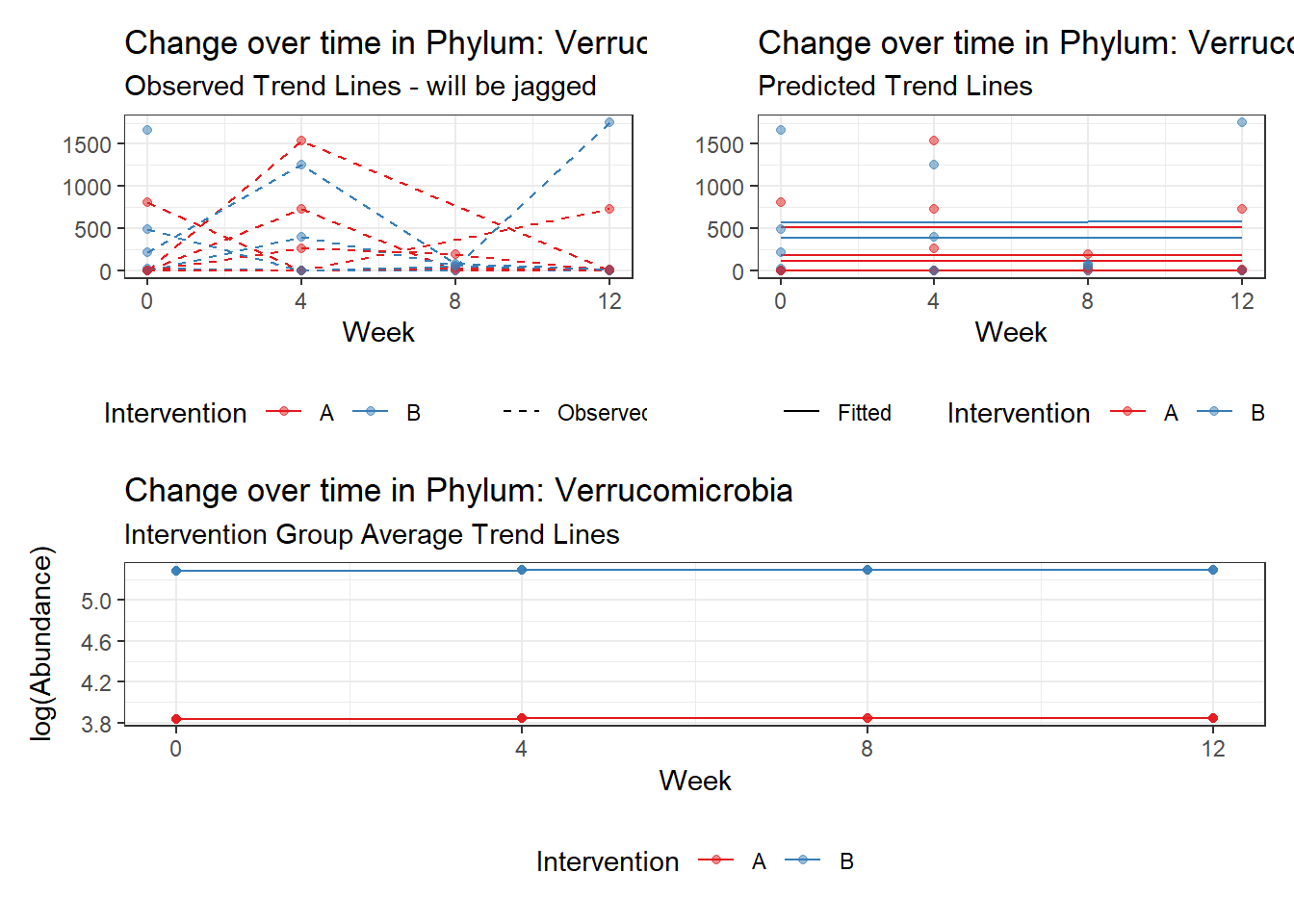
Model: Verrucomicrobia ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x0000000049981218>
Link: poisson
Intraclass Correlation (ICC): 0.8508851

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 3.83630 0.99216 3.86662 0.00011
intB Verrucomicrobia 1.44885 1.45824 0.99356 0.32044
time Verrucomicrobia 0.00500 0.00920 0.54381 0.58657
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3888
Significance test not available for random effectsphylum.fit4b <- glmm_microbiome(mydata=microbiome_data,model.number=4,
taxa.level="Phylum", link="negbinom",
model="1 + intB + time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinom
Intraclass Correlation (ICC): 0.3000096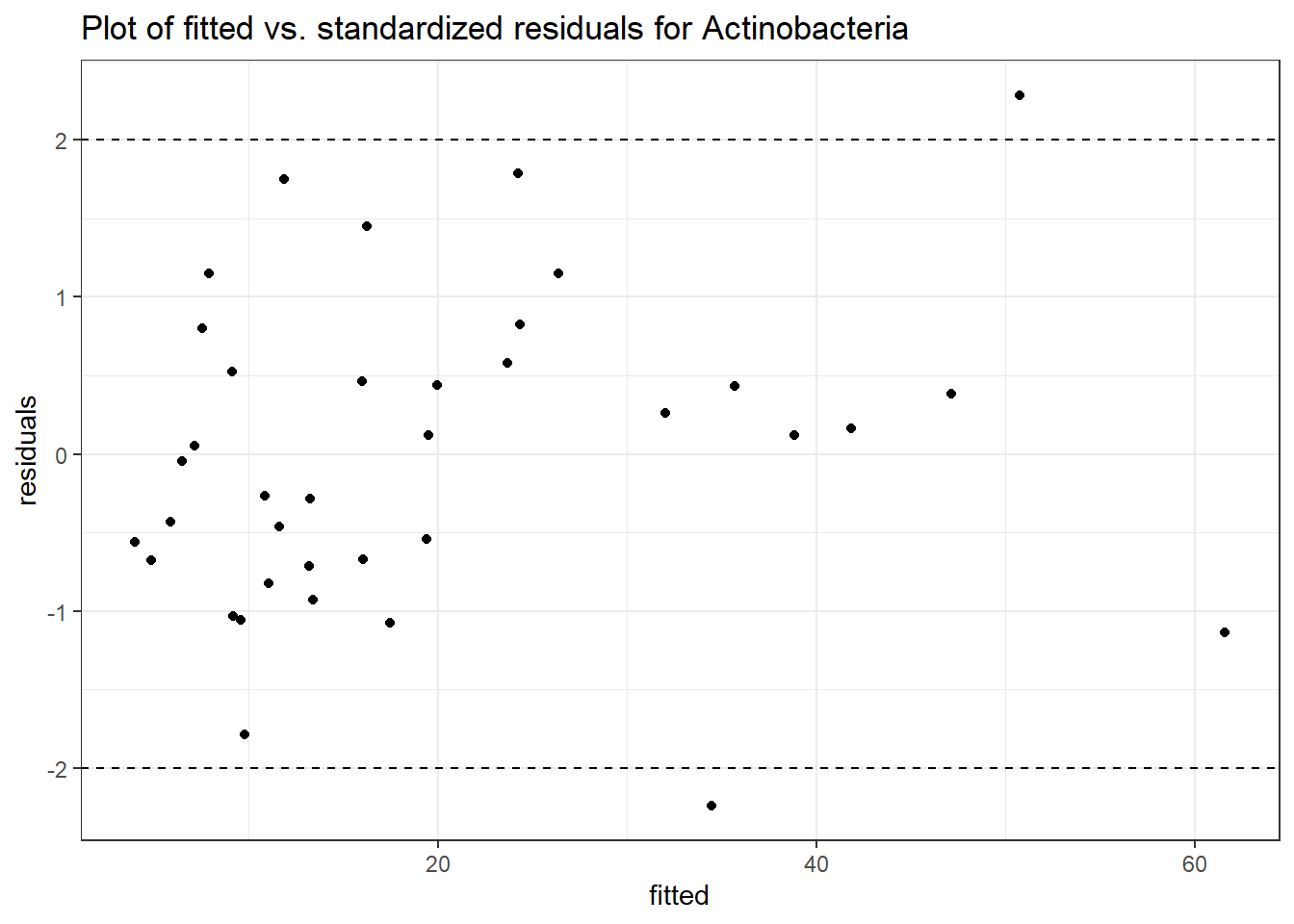
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.12141 0.46053 4.60642 0.00000
intB Actinobacteria 0.87112 0.56308 1.54706 0.12185
time Actinobacteria 0.19350 0.18685 1.03558 0.30040
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.65467
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinom
Intraclass Correlation (ICC): 0.1360569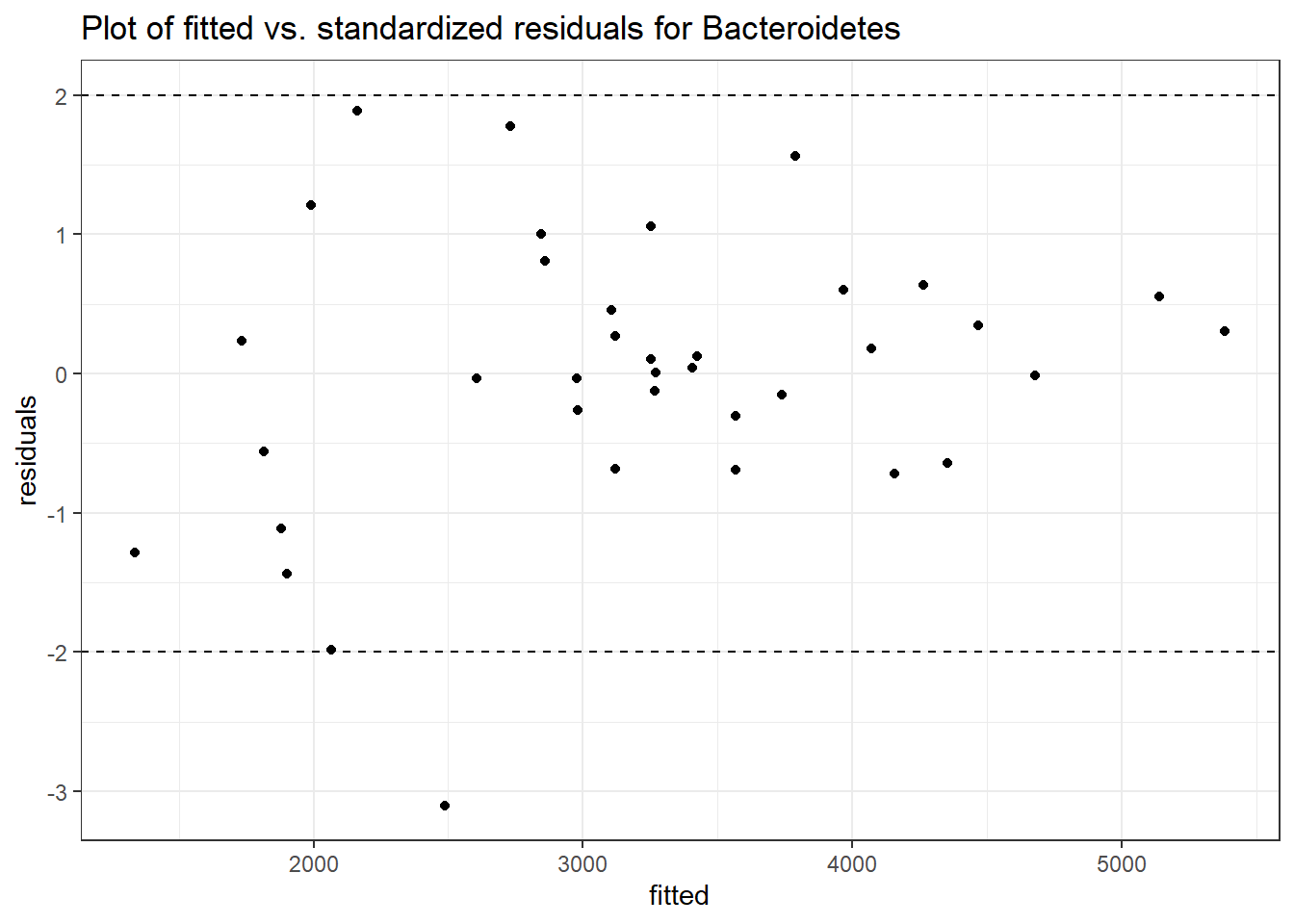
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.15117 0.17650 46.18176 0.00000
intB Bacteroidetes -0.23442 0.25640 -0.91426 0.36058
time Bacteroidetes -0.04643 0.03469 -1.33847 0.18074
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.39684
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 6.414827e-14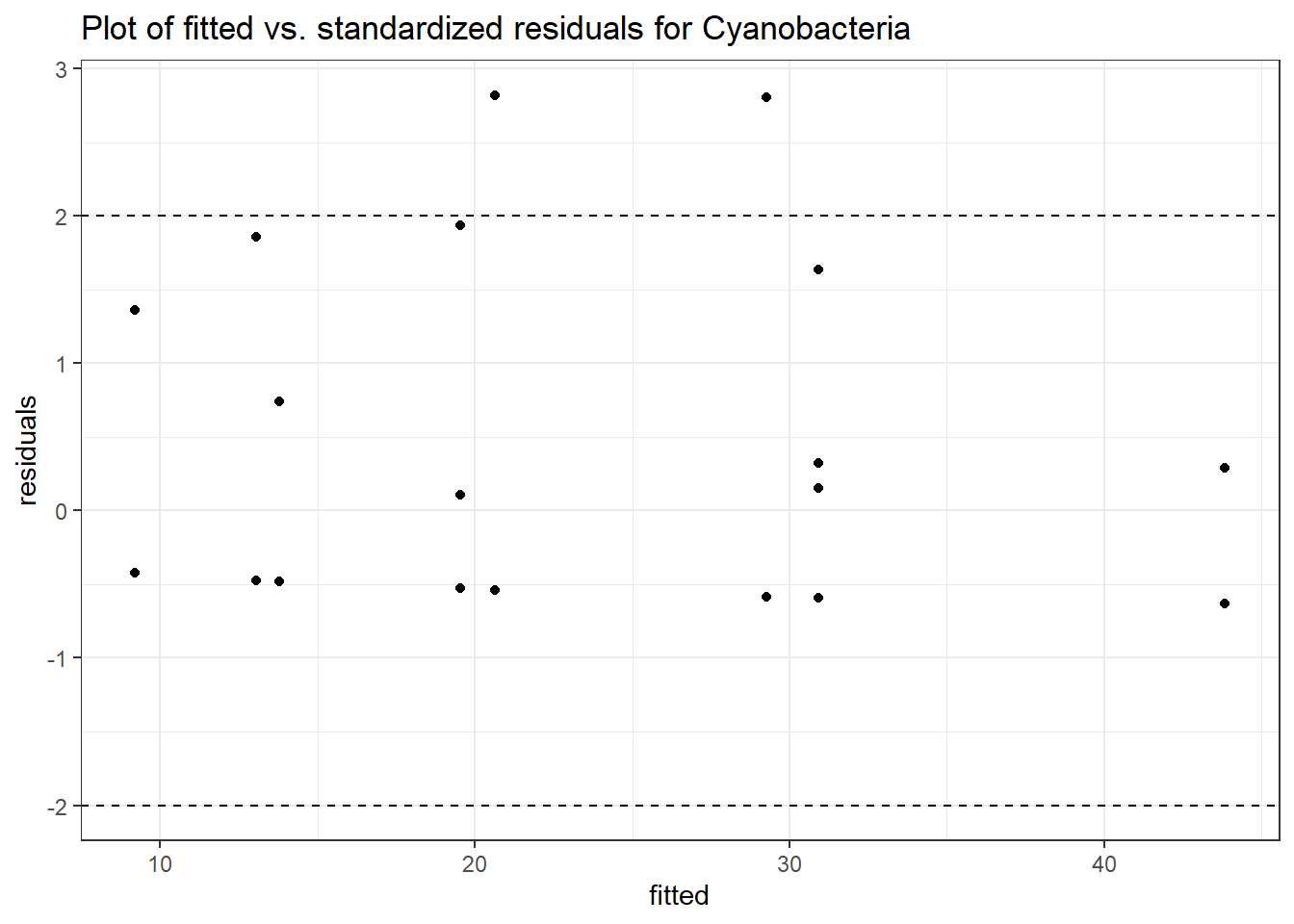
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 3.78059 1.61294 2.34392 0.01908
intB Cyanobacteria -0.34917 1.41330 -0.24706 0.80486
time Cyanobacteria -0.40424 0.81506 -0.49597 0.61992
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.5328e-07
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinomboundary (singular) fit: see ?isSingular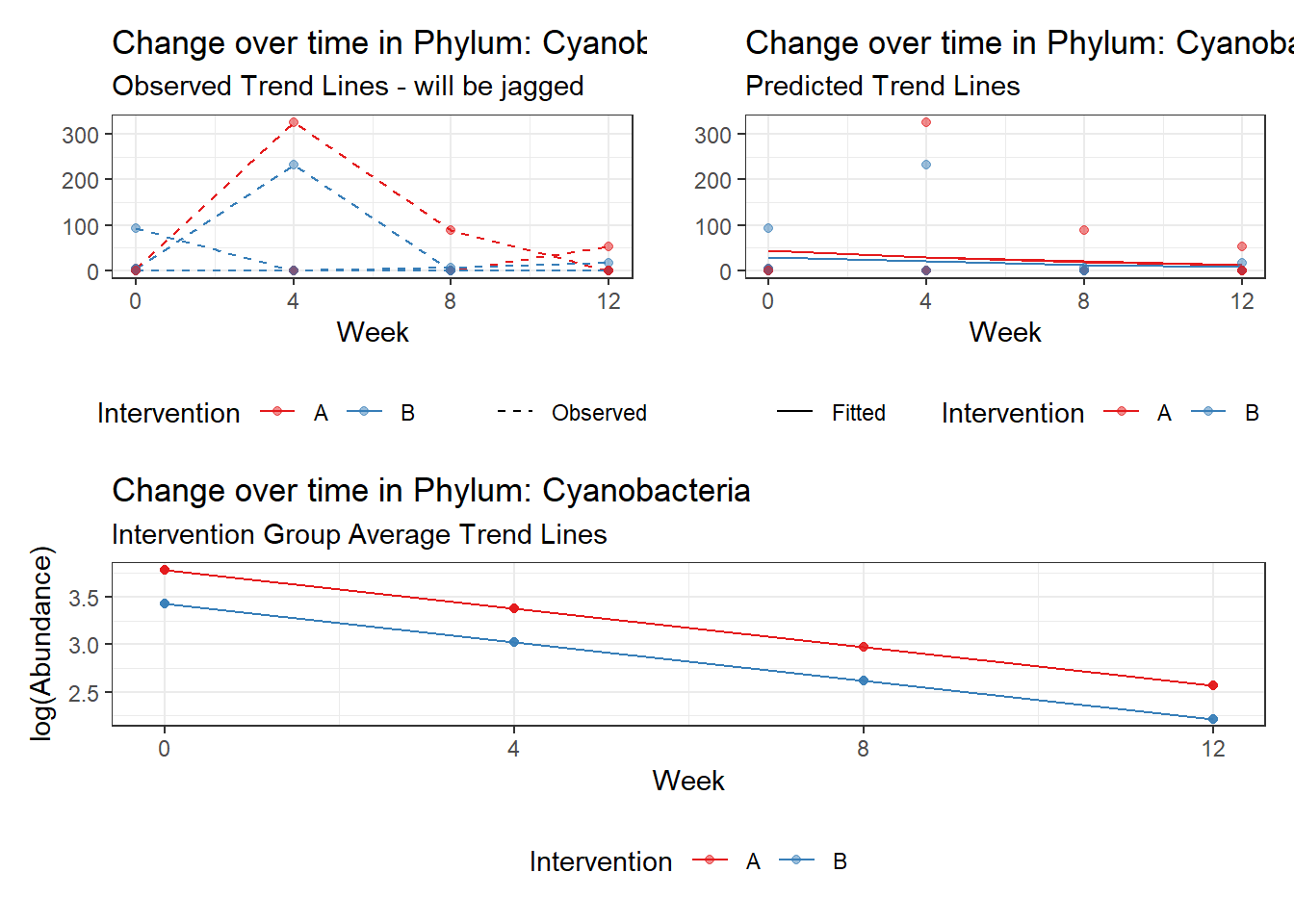
Intraclass Correlation (ICC): 3.391275e-13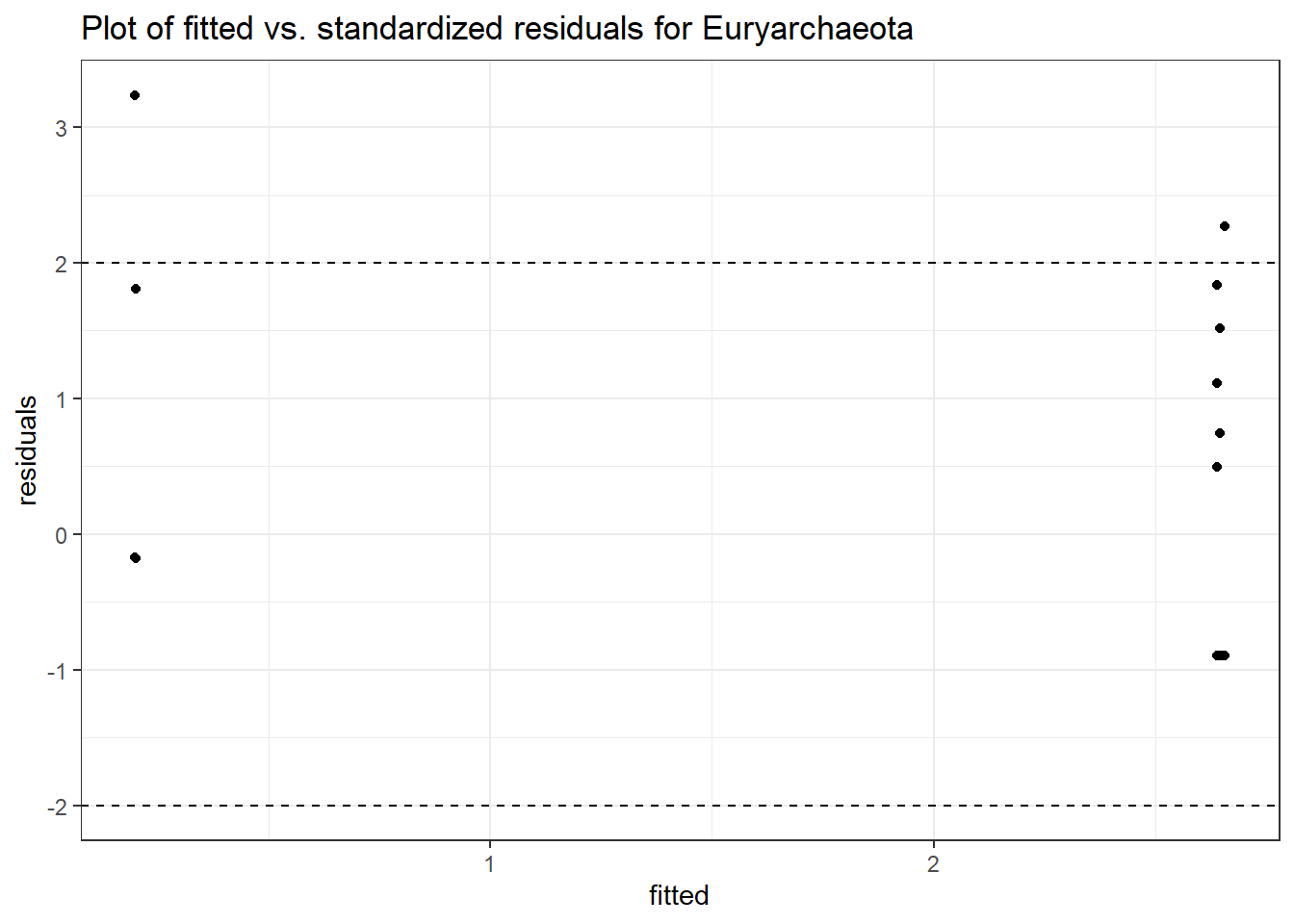

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -1.61269 0.96423 -1.67251 0.09442
intB Euryarchaeota 2.58316 0.99611 2.59325 0.00951
time Euryarchaeota 0.00226 0.41232 0.00548 0.99562
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 5.8235e-07
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinom
Intraclass Correlation (ICC): 0.1178534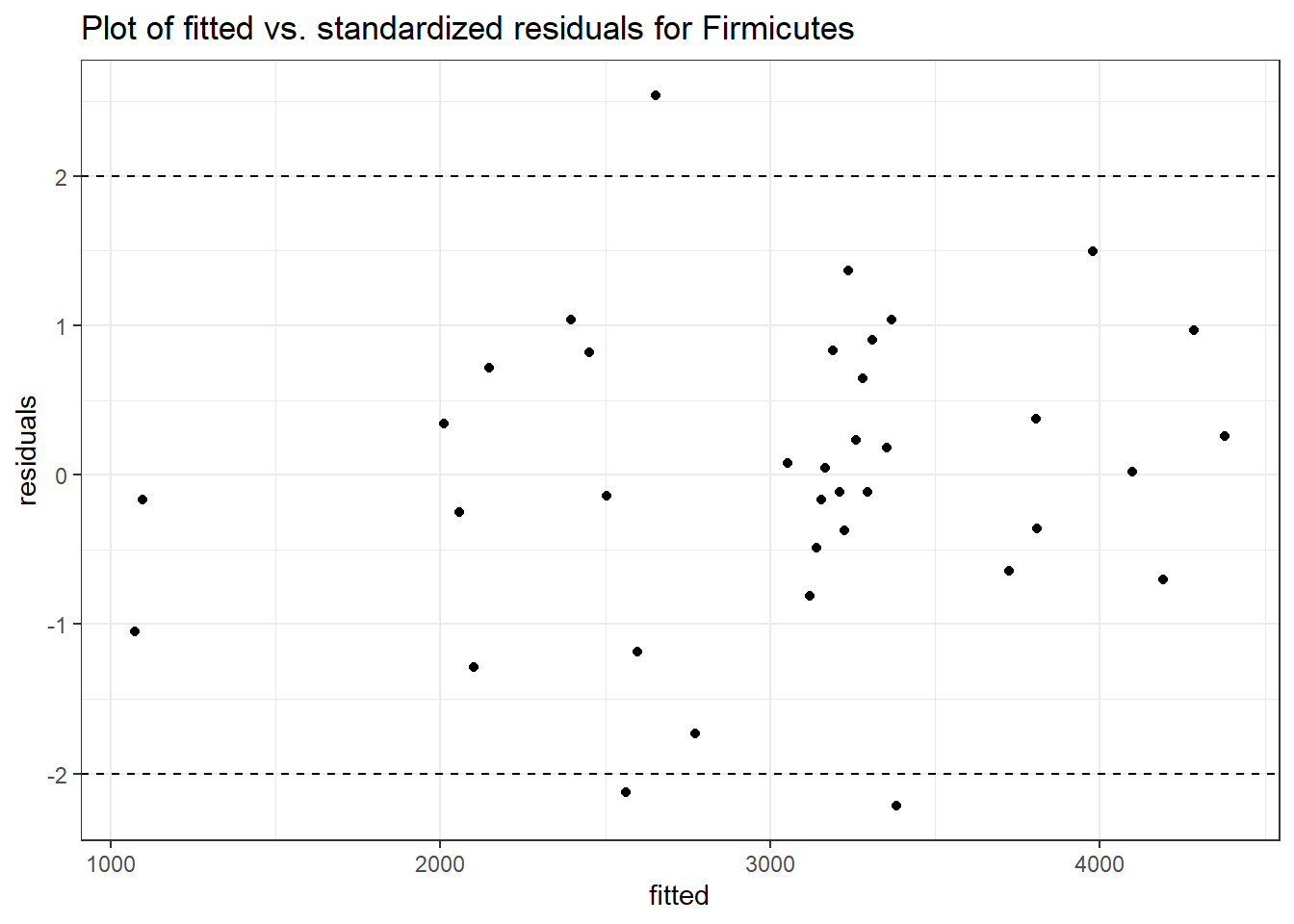
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.86748 0.15694 50.13155 0.00000
intB Firmicutes 0.13328 0.22878 0.58257 0.56018
time Firmicutes 0.02203 0.02408 0.91491 0.36024
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.36551
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0118533 (tol = 0.002, component 1)
Intraclass Correlation (ICC): 0.8141549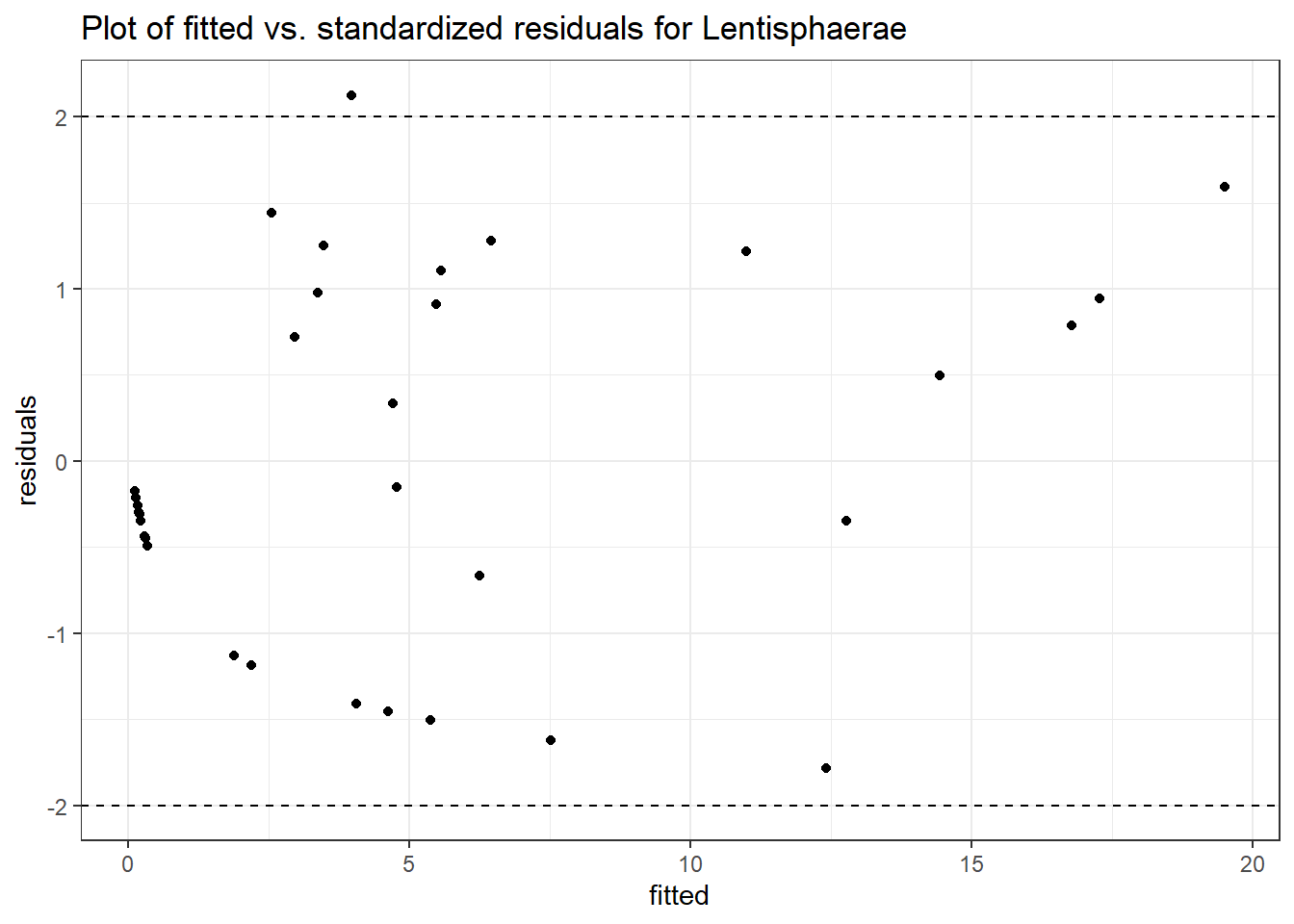
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.38123 1.12249 0.33962 0.73414
intB Lentisphaerae -0.38367 1.48971 -0.25755 0.79676
time Lentisphaerae 0.15076 0.28310 0.53254 0.59435
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.093
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinom
Intraclass Correlation (ICC): 0.02306262
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.89356 0.18279 32.24277 0.00000
intB Proteobacteria -0.21404 0.22838 -0.93720 0.34866
time Proteobacteria -0.04562 0.09209 -0.49541 0.62031
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.15365
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinomboundary (singular) fit: see ?isSingular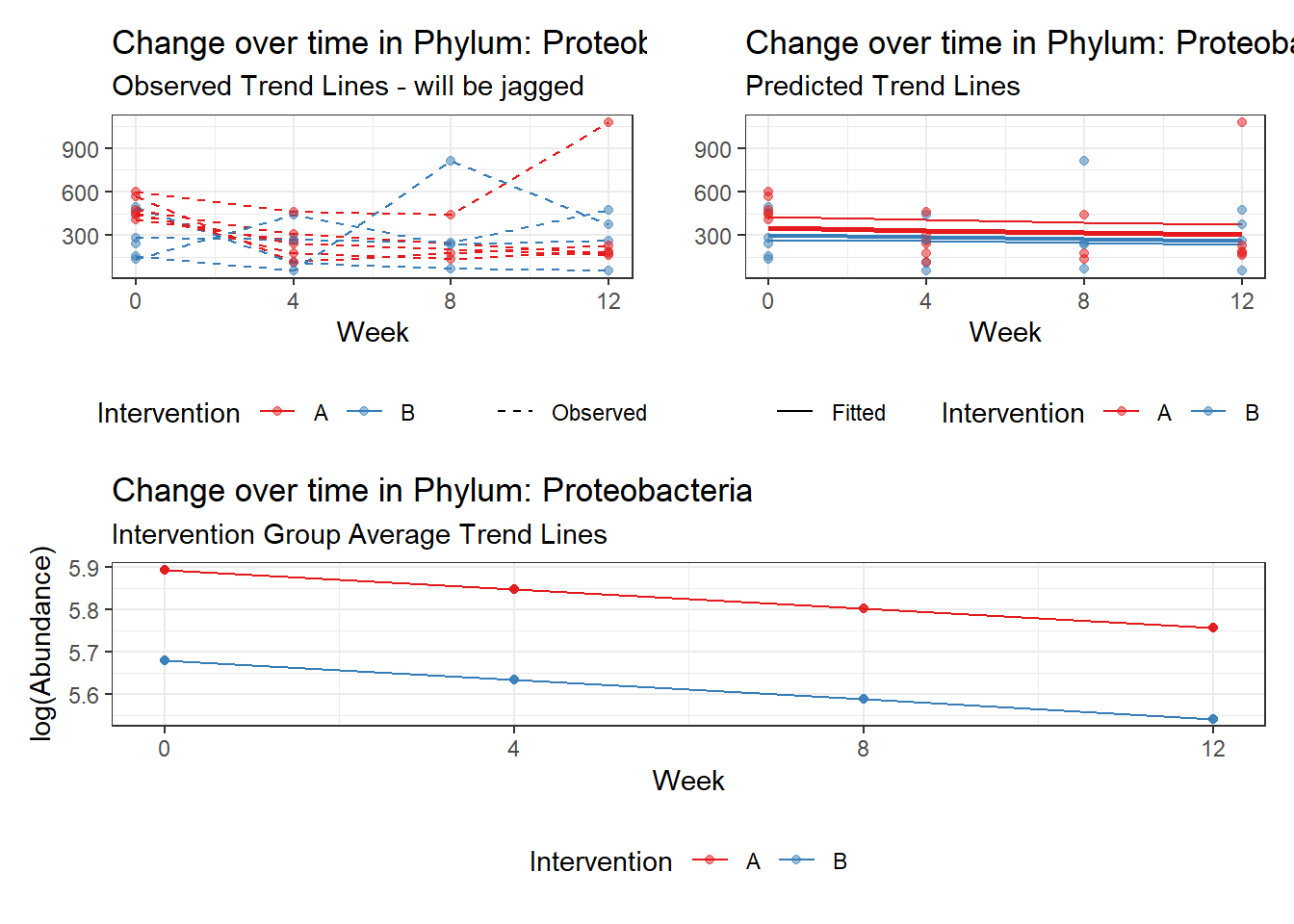
Intraclass Correlation (ICC): 2.145298e-11
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 2.56067 0.92499 2.76831 0.00563
intB Tenericutes 0.02650 1.04348 0.02540 0.97974
time Tenericutes 0.70752 0.45338 1.56054 0.11863
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.6317e-06
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003131b0a8>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced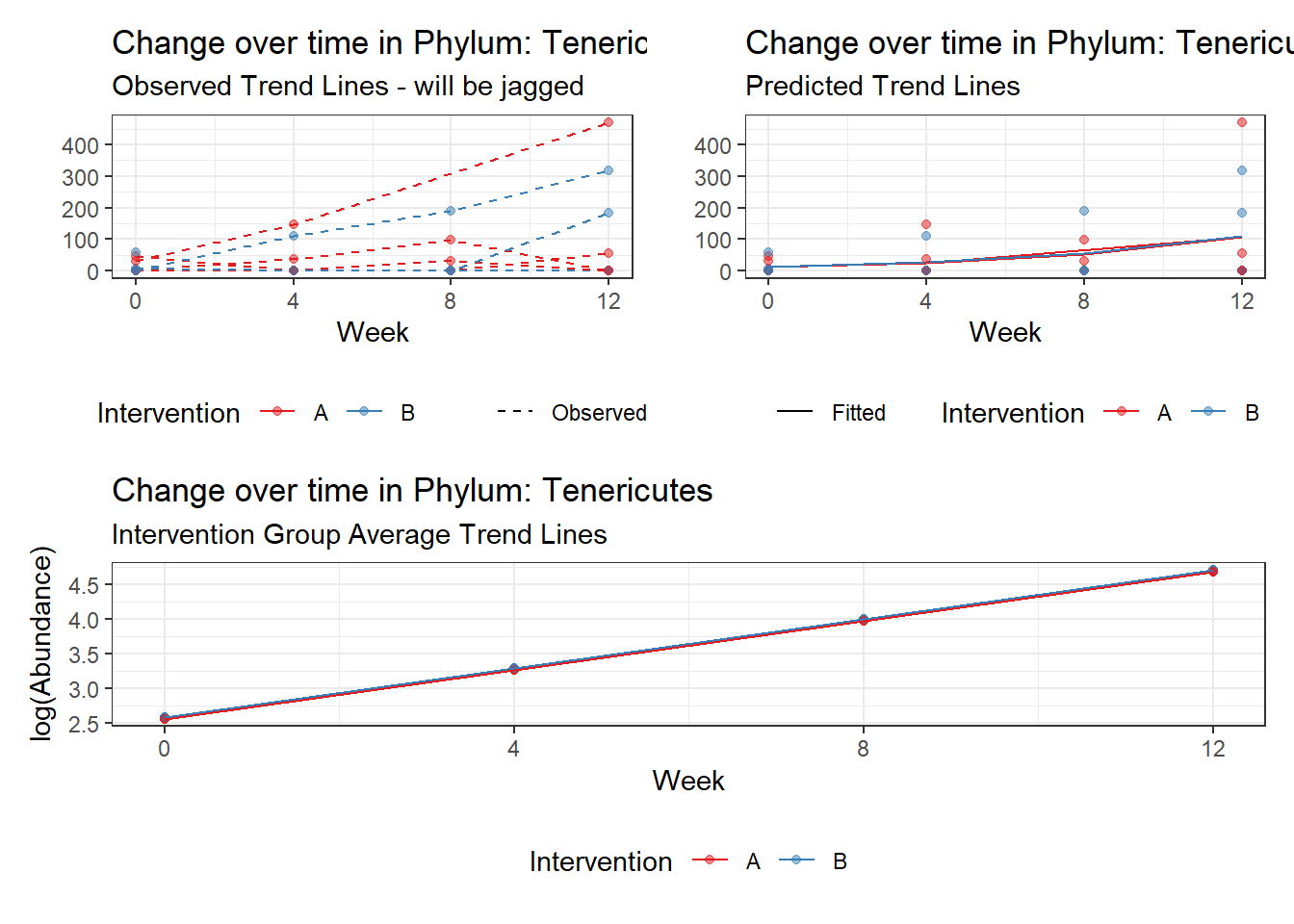
Intraclass Correlation (ICC): 0.8511595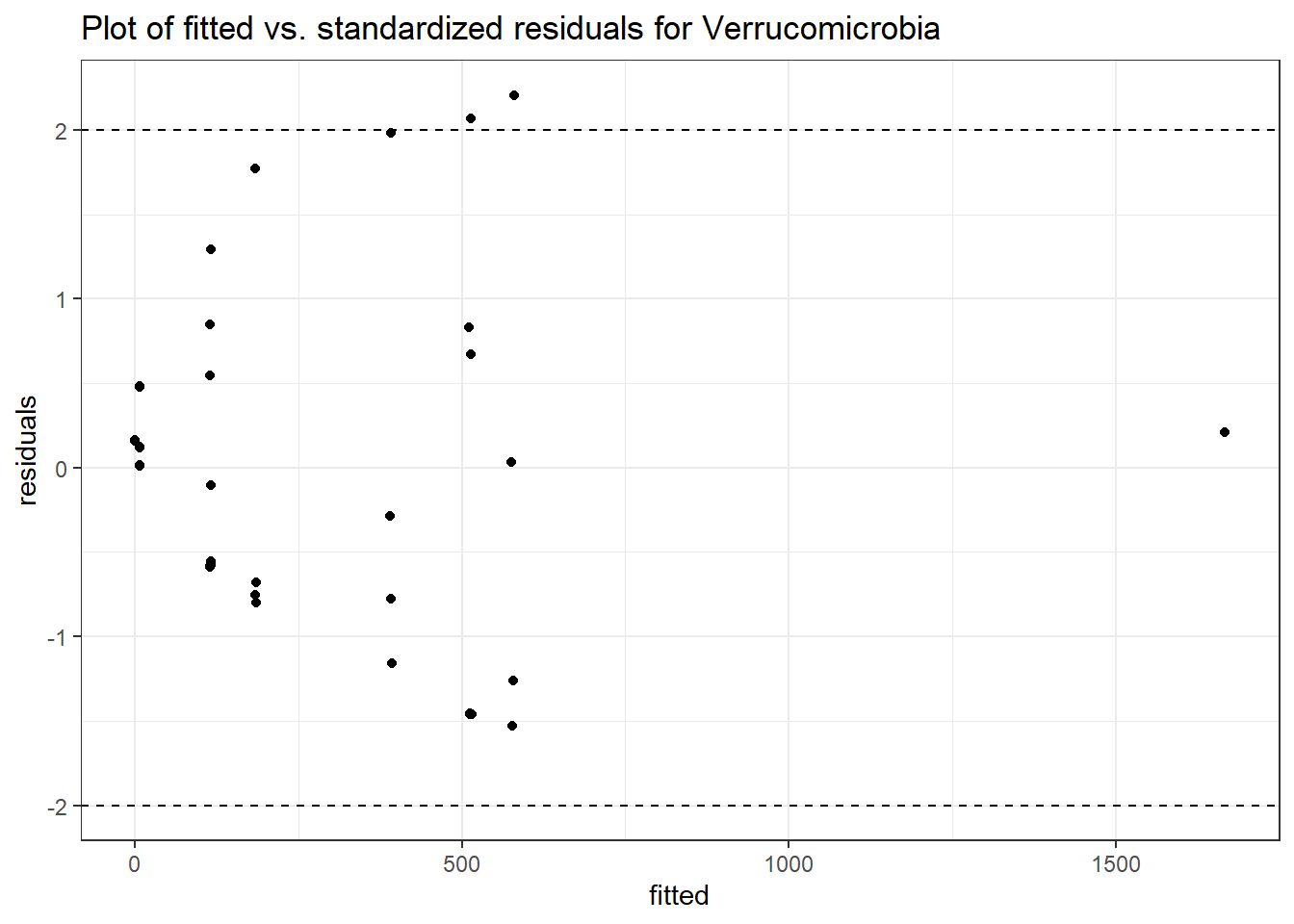
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 3.83782 0.99389 3.86142 0.00011
intB Verrucomicrobia 1.45103 1.46178 0.99264 0.32088
time Verrucomicrobia 0.00189 0.00938 0.20199 0.83993
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3914
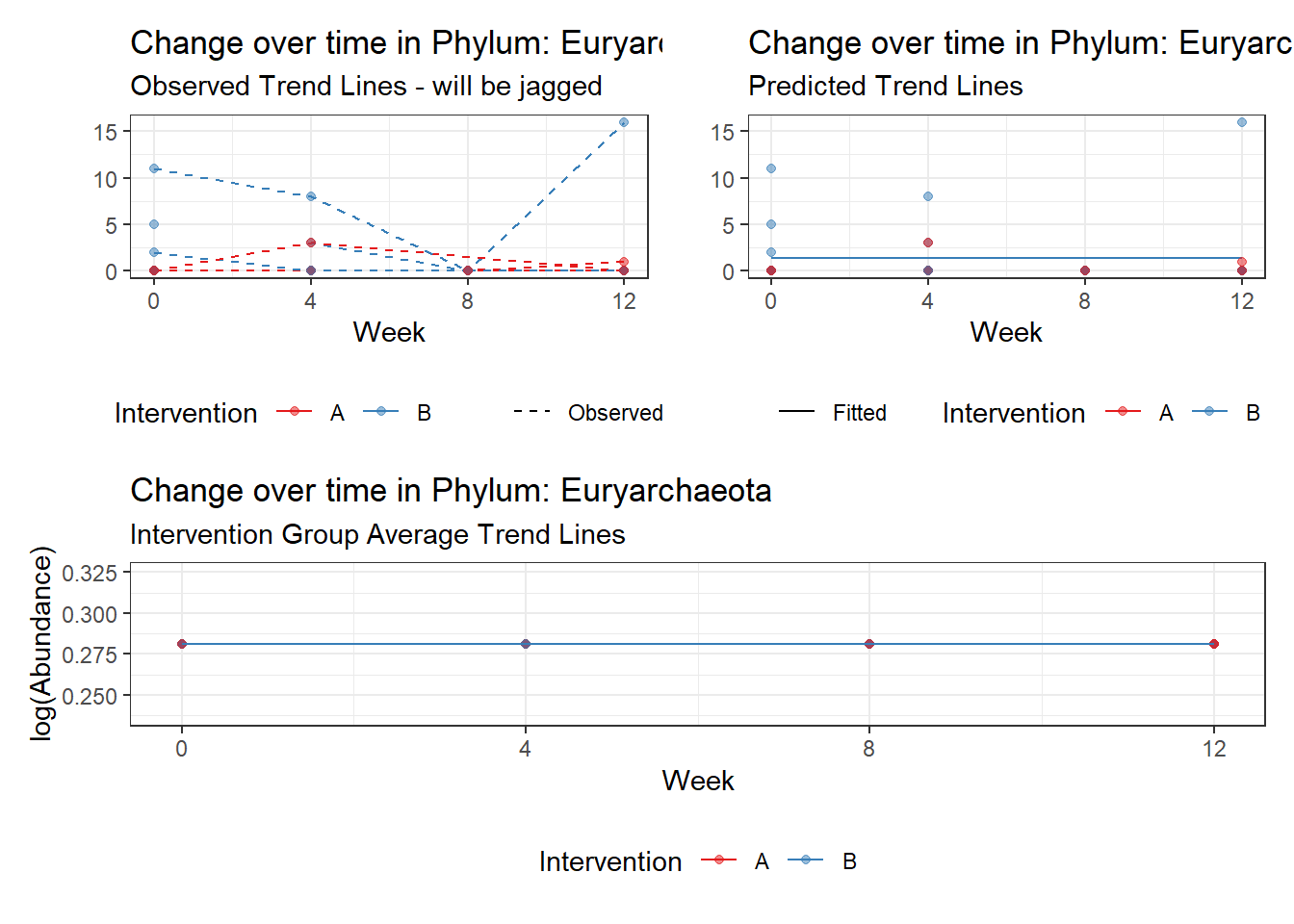
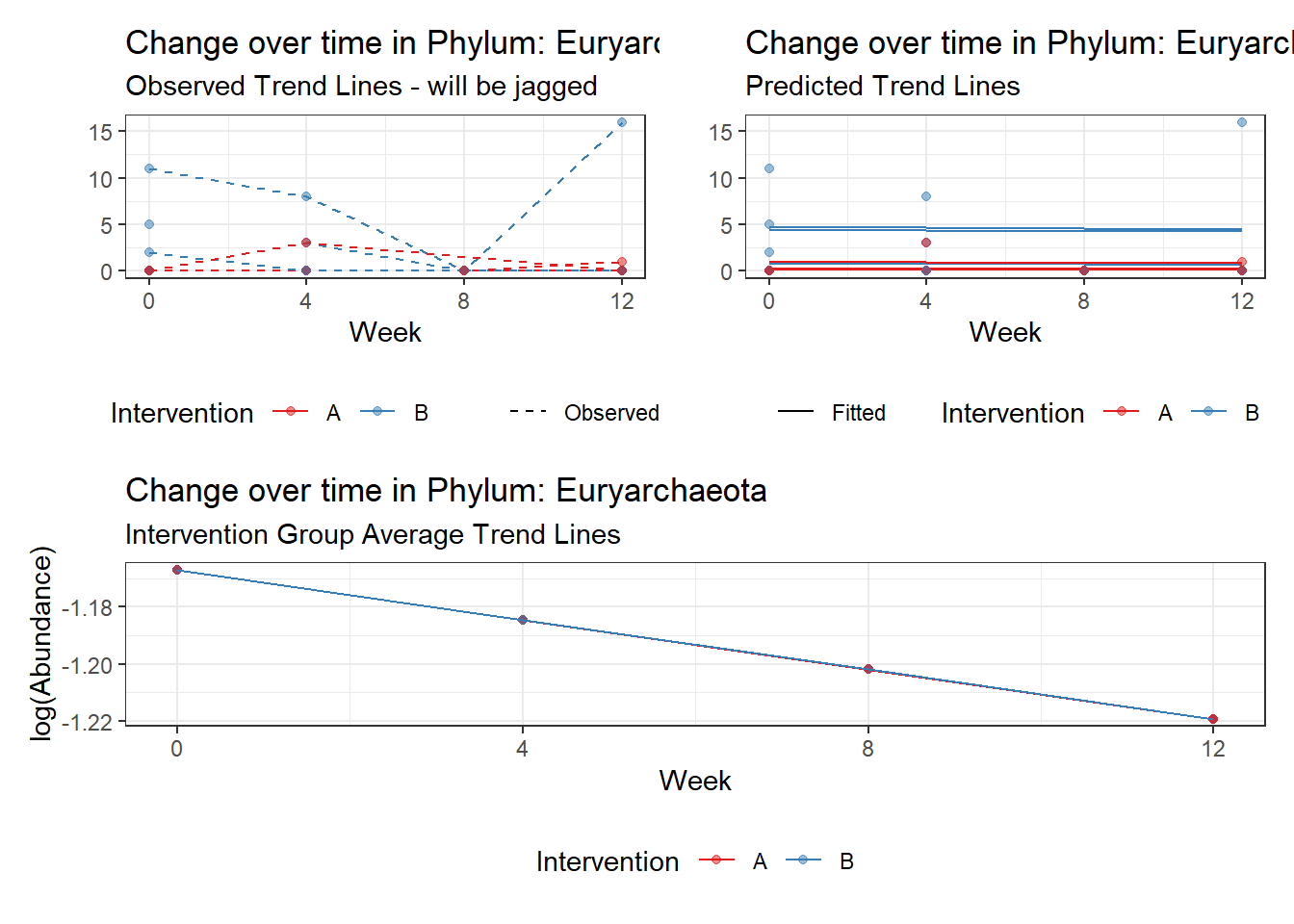
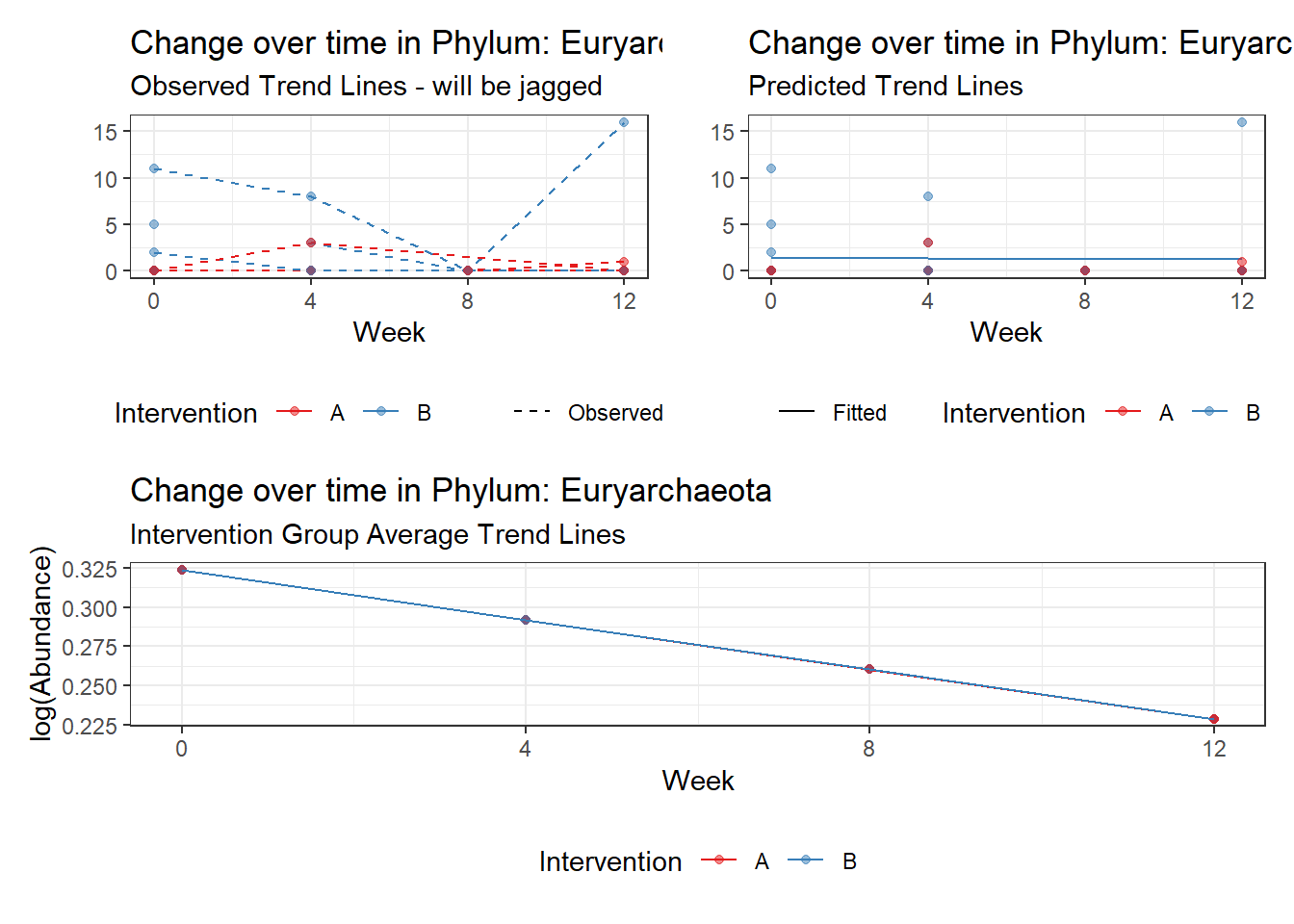
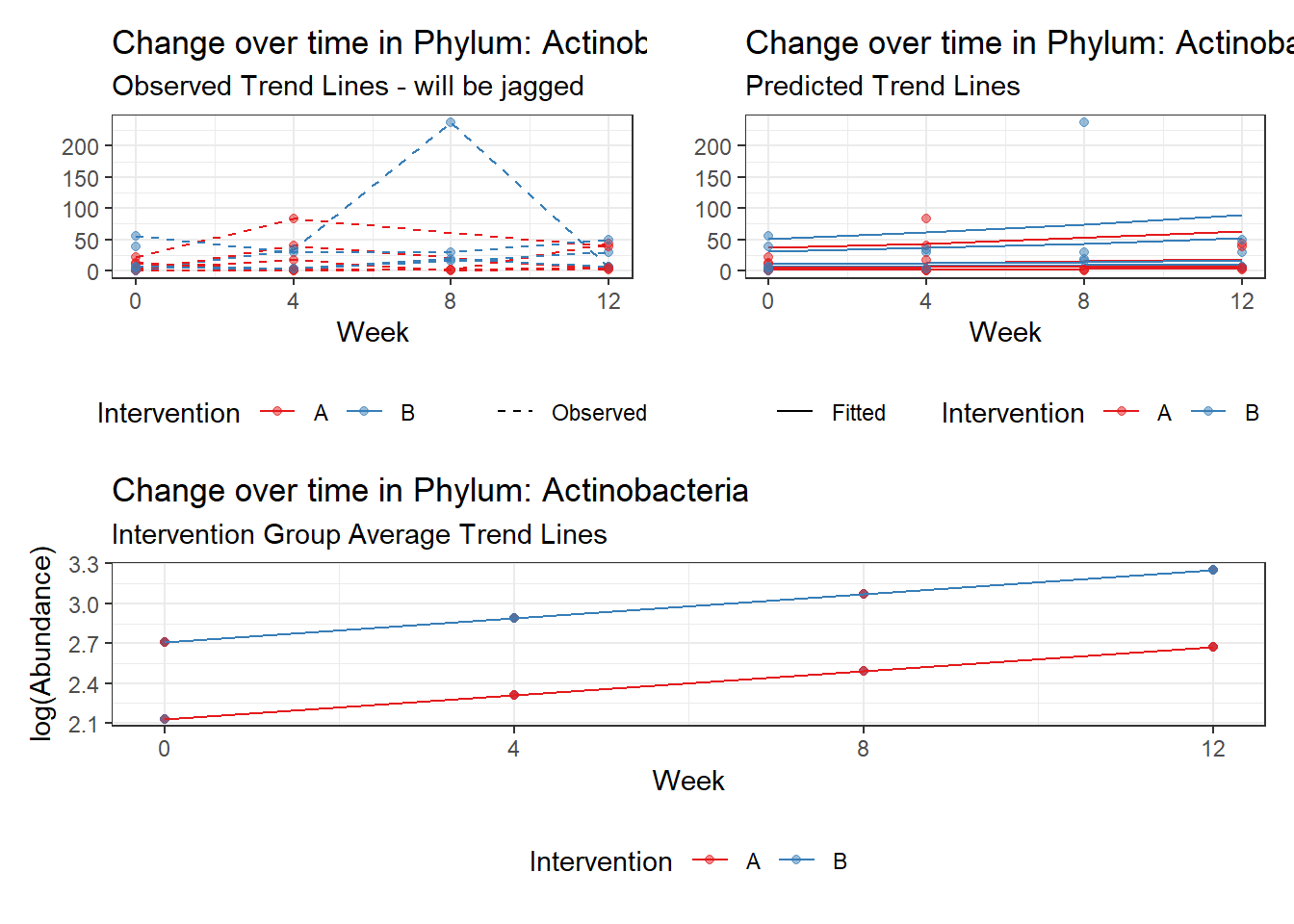
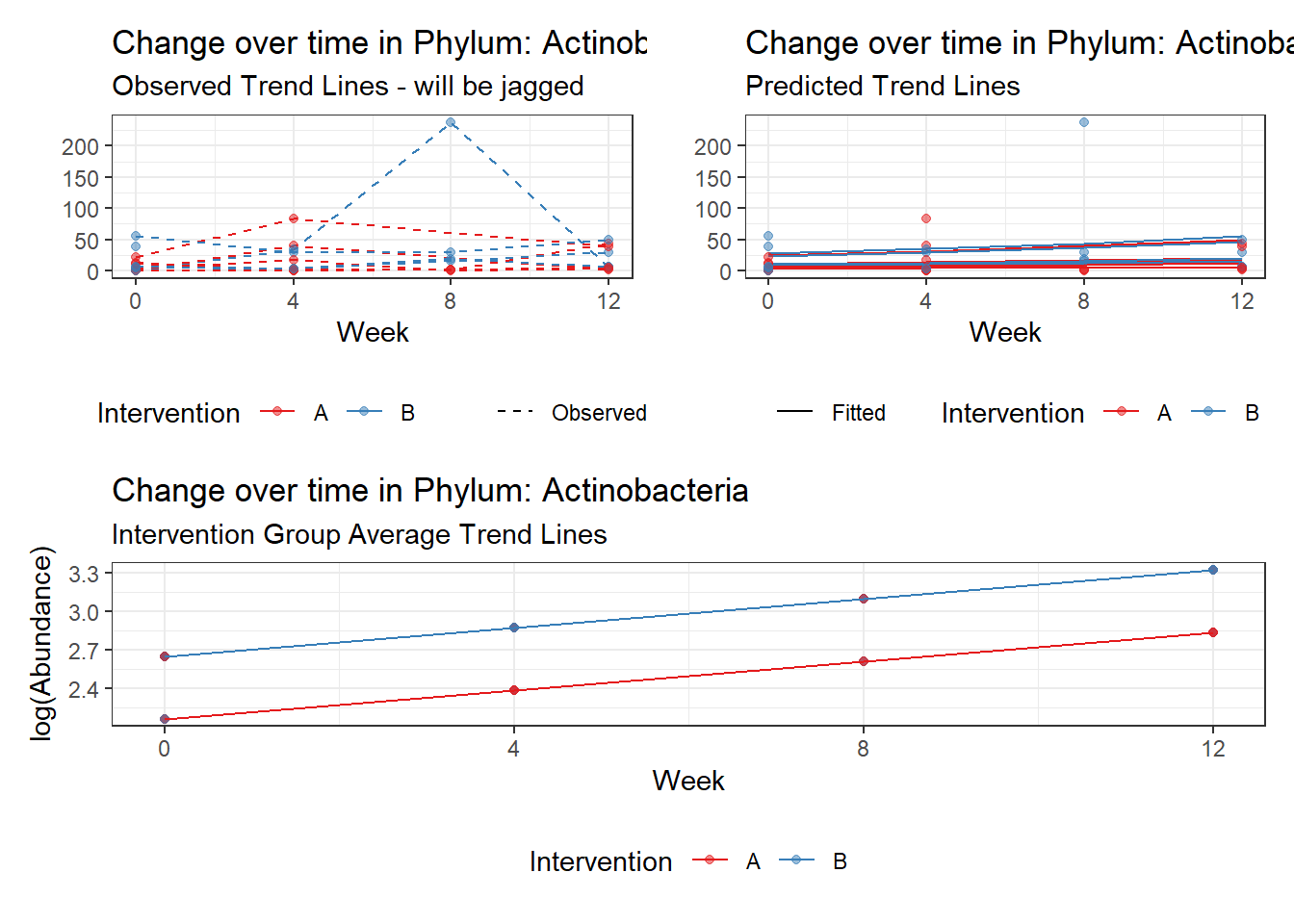
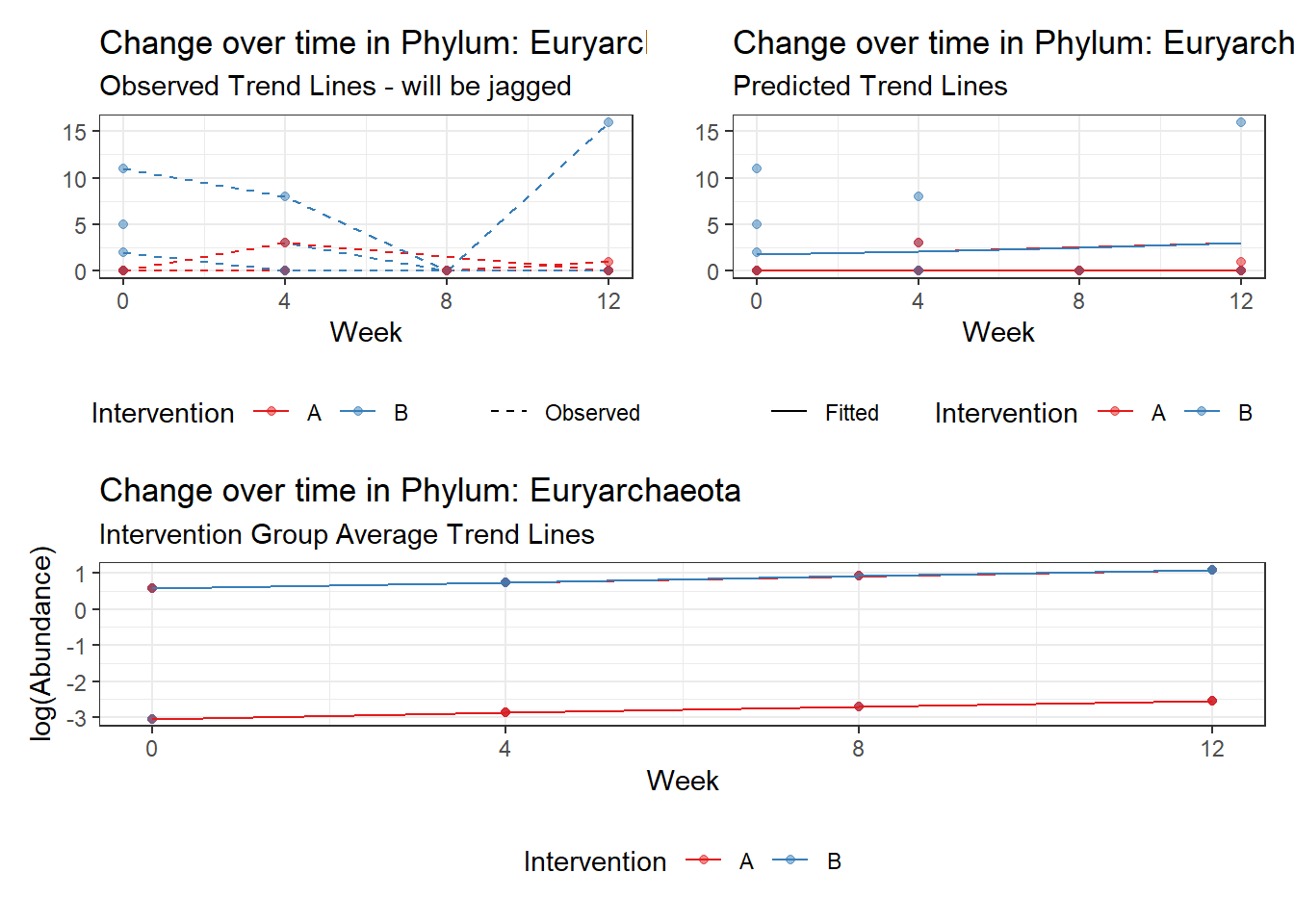
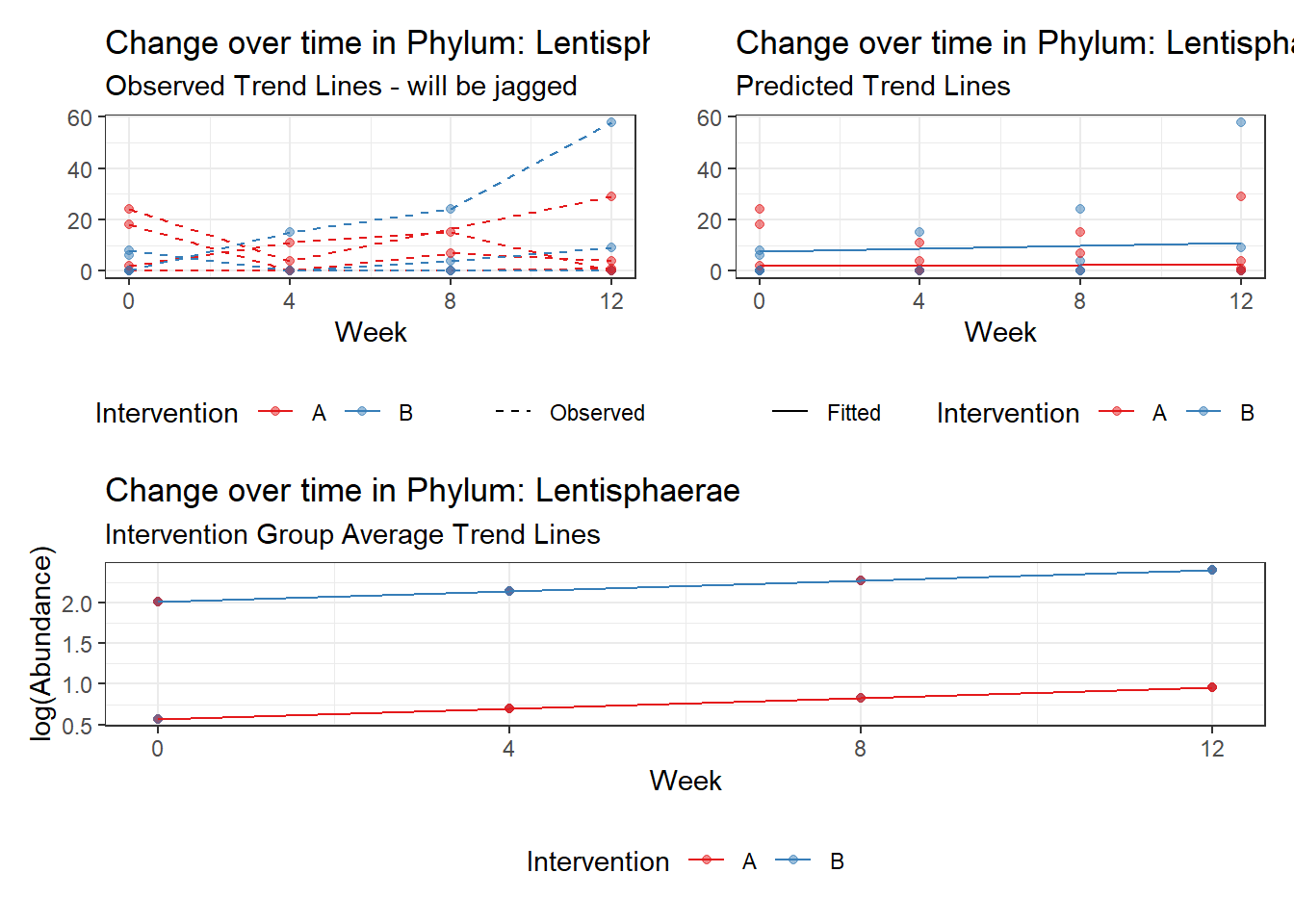
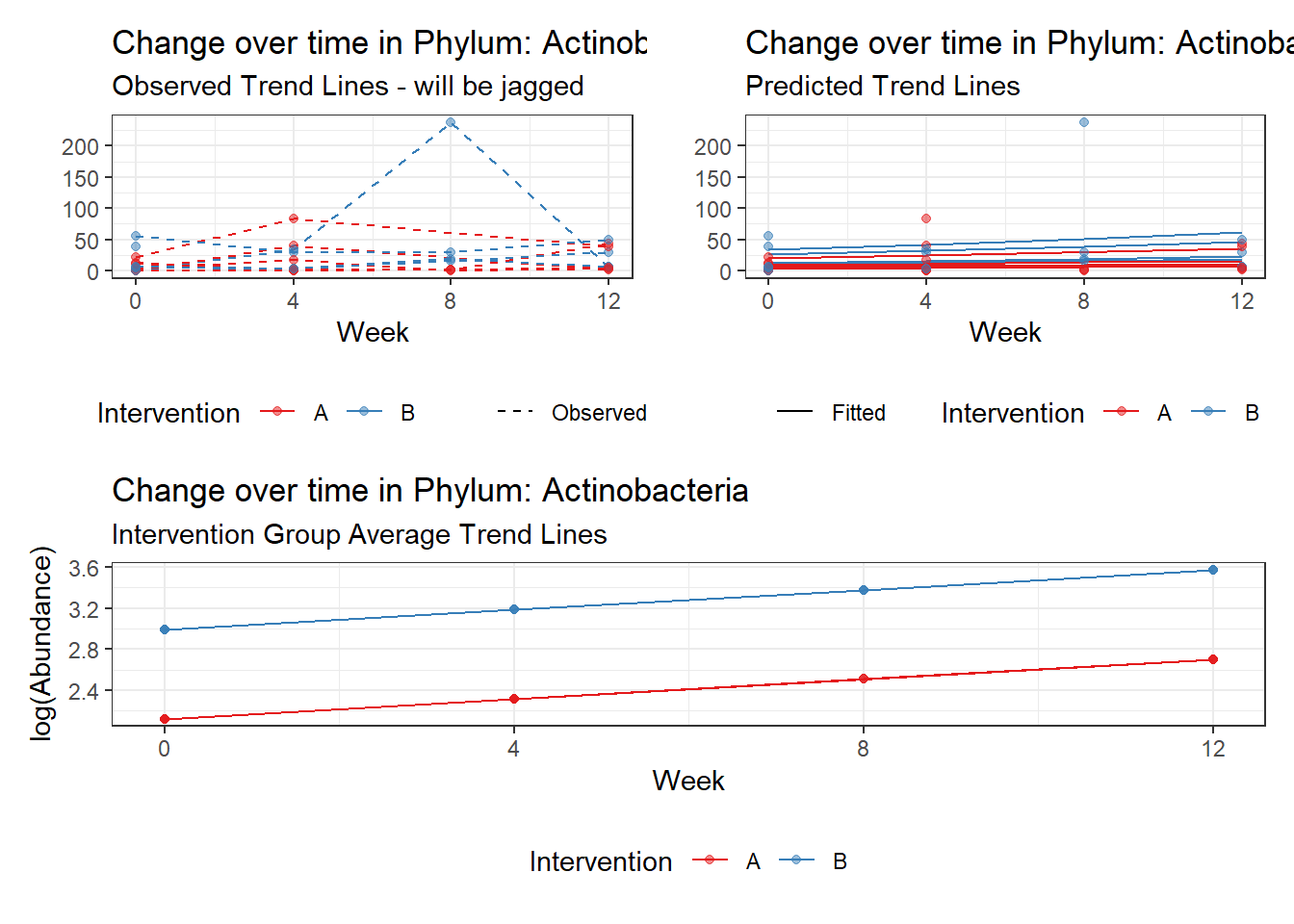
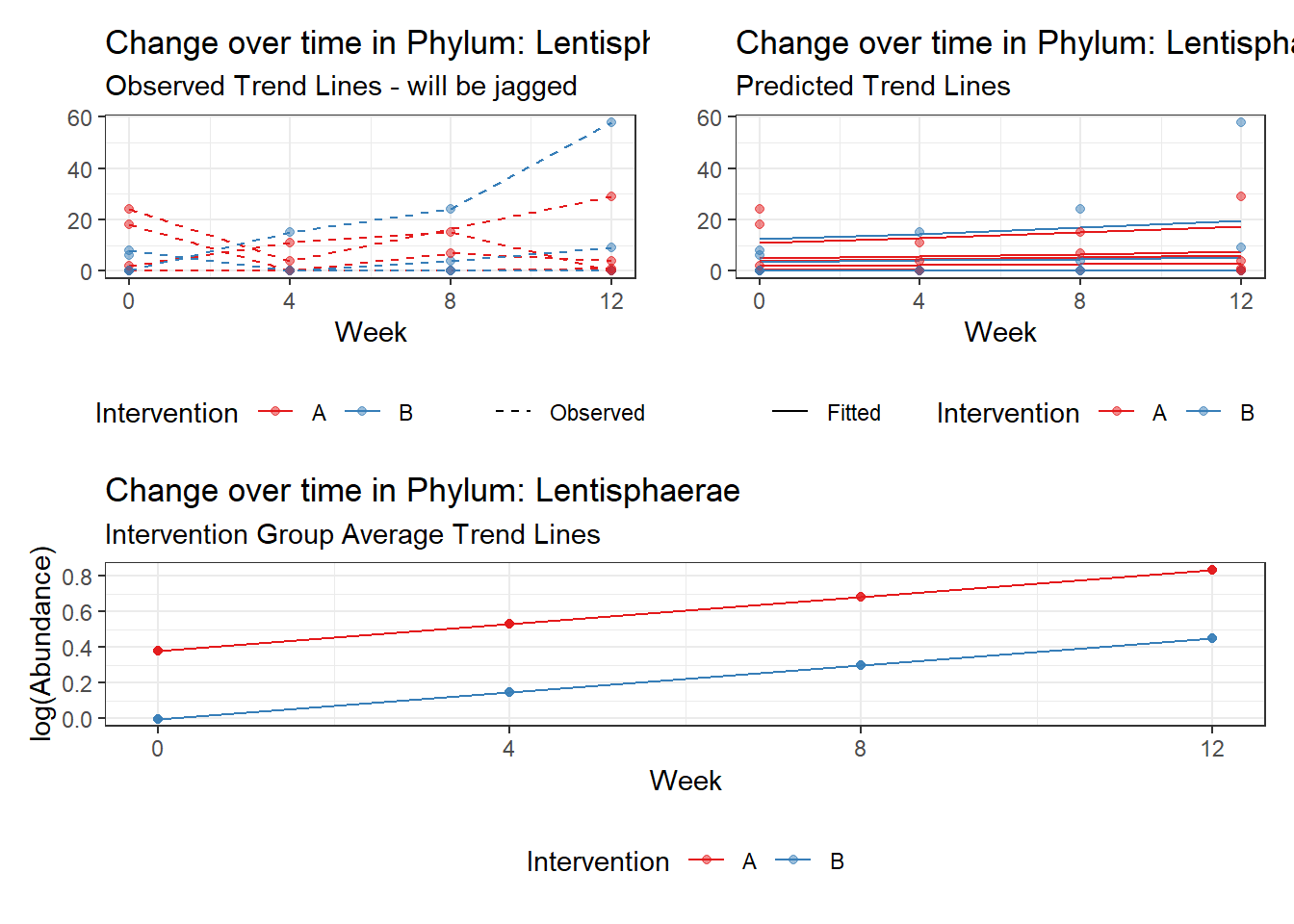
Significance test not available for random effectsControlling for the effect of time, the intervention resulted in a significant increase in abundance of Phylums: Euryarchaeota. This is in contrast to the effect of time, which was negatively associated with abundance. So, abundance of Euryarchaeota increased in intervention group B but not in group A.
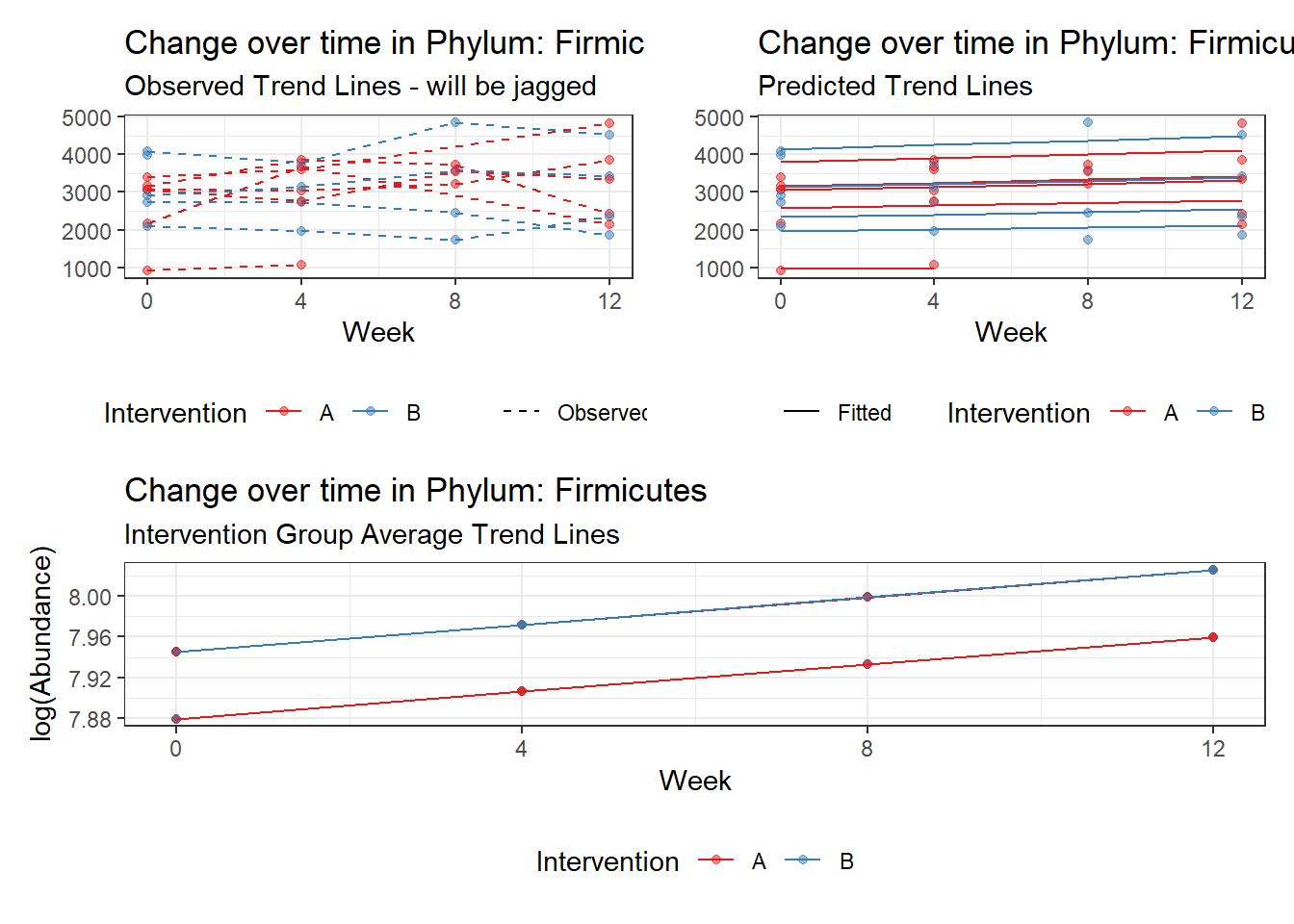
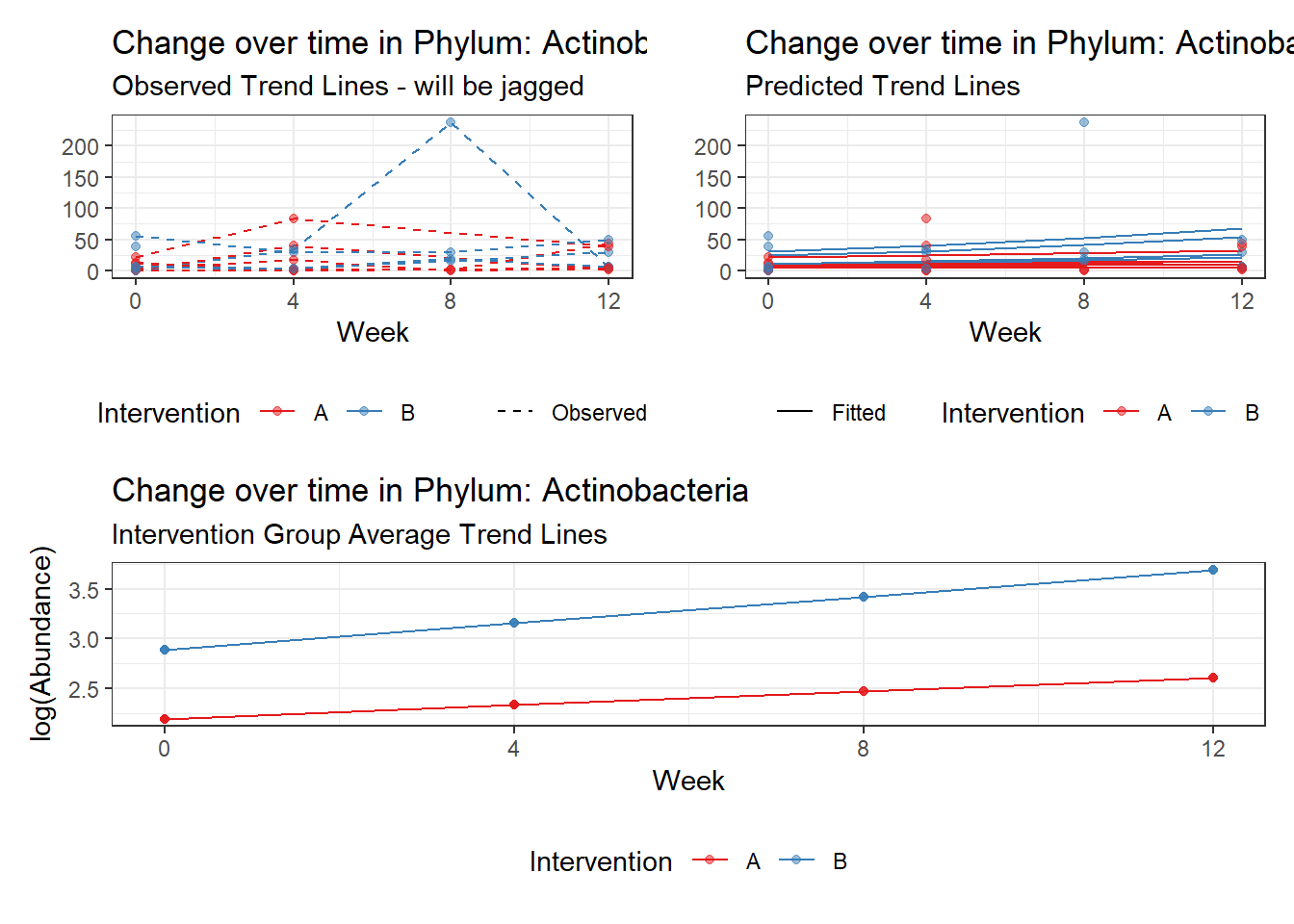
Other interesting but not significant results. The effect of the intervention was in the opposite direction (positive) compared to the effect of time (negative) is Phylums: Actinobacteria, Firmicutes, Lentisphaerae, and Tenericutes.
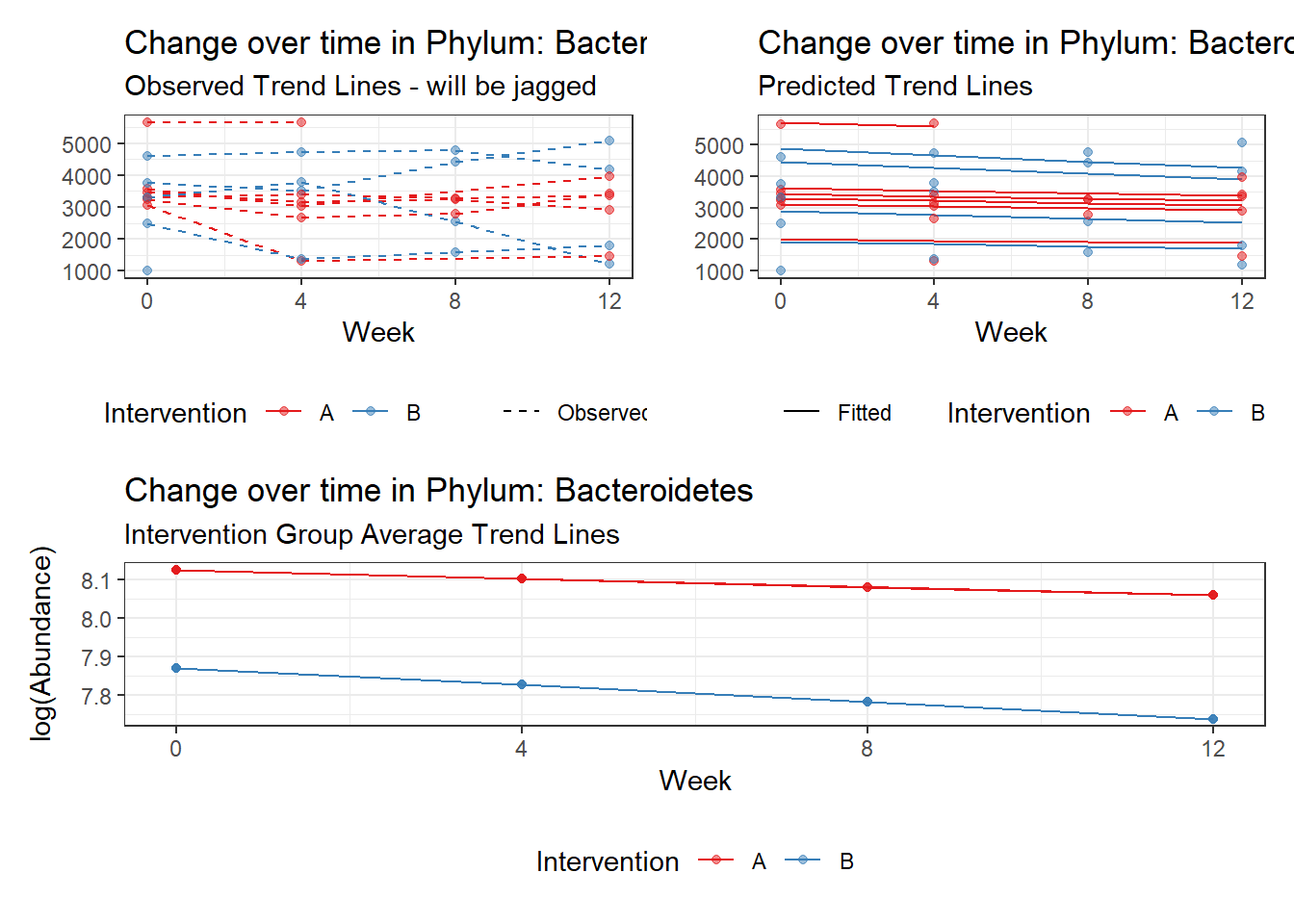
Covariate(s): Time by intervention interaction
Here, the model tests whether the effect of the intervention is constant across time or does the effect of the intervention get stronger (or weaker) at time progress. And remember, time is coded 0, 1, 2, 3, so a 1 unit increase in time corresponds to 1 month or 4 weeks of the intervention.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Intervention)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
It could be that the intervention is directly related to time so included a average effect of time through the main effect effect is just adding more parameters and is not informative. If the effect of the intervention is still essentially zero, we will re-estimate the model without the main effect.
phylum.fit5 <- glmm_microbiome(mydata=microbiome_data,model.number=5,
taxa.level="Phylum", link="poisson",
model="1 + intB + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.436752
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.17672 0.37536 5.79901 0.00000
intB Actinobacteria 0.78150 0.54991 1.42113 0.15528
time Actinobacteria 0.09795 0.04867 2.01258 0.04416
intB:time Actinobacteria 0.13414 0.06271 2.13884 0.03245
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.88058
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.1703446
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.12428 0.18509 43.89474 0.00000
intB Bacteroidetes -0.25420 0.27458 -0.92577 0.35456
time Bacteroidetes -0.02187 0.00363 -6.02179 0.00000
intB:time Bacteroidetes -0.02236 0.00532 -4.20690 0.00003
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.45312
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.9437787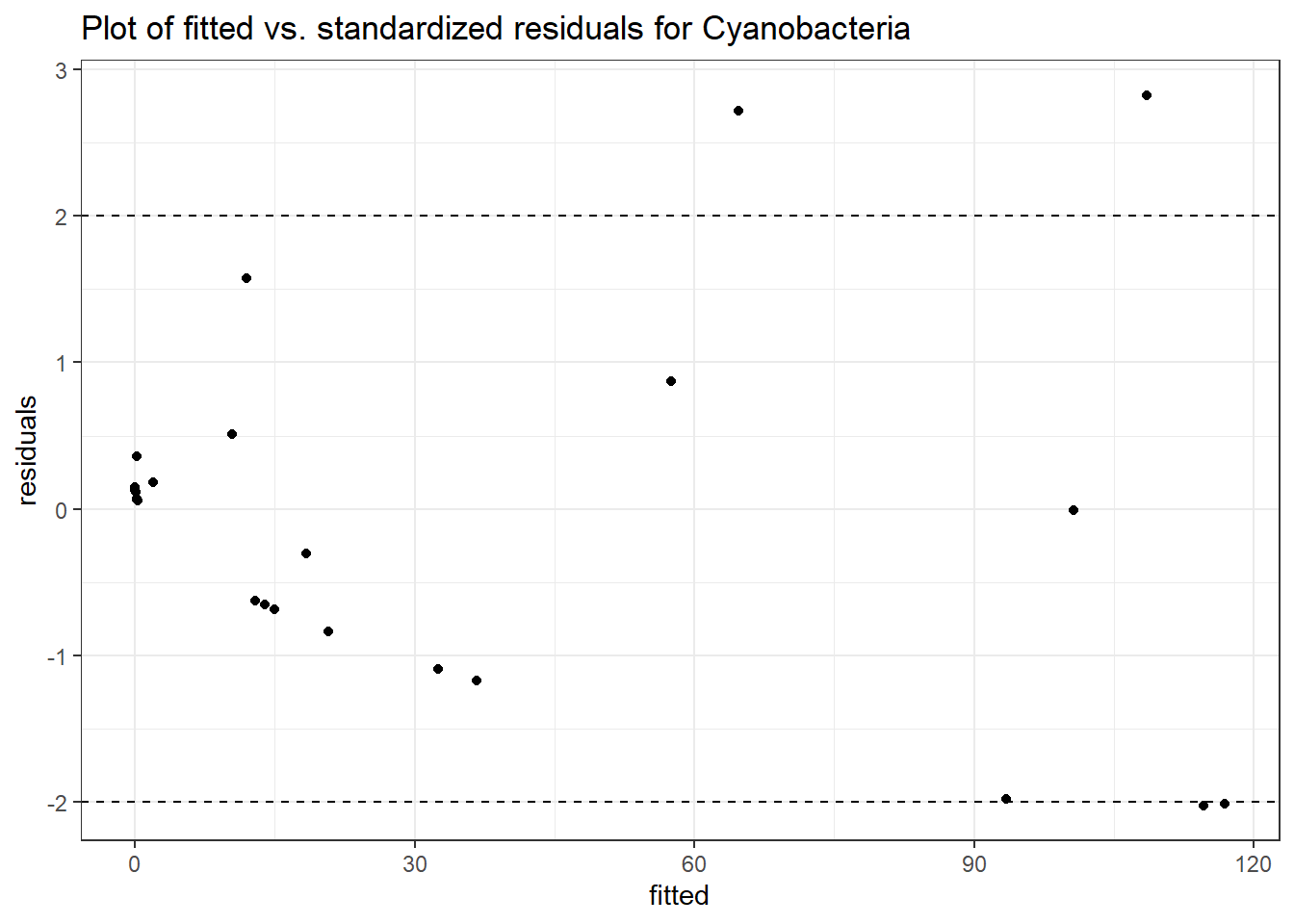

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -1.65226 2.22499 -0.74259 0.45773
intB Cyanobacteria 1.82089 2.84741 0.63949 0.52250
time Cyanobacteria -0.07461 0.04114 -1.81363 0.06973
intB:time Cyanobacteria -0.49582 0.06785 -7.30780 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.0972
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.6283401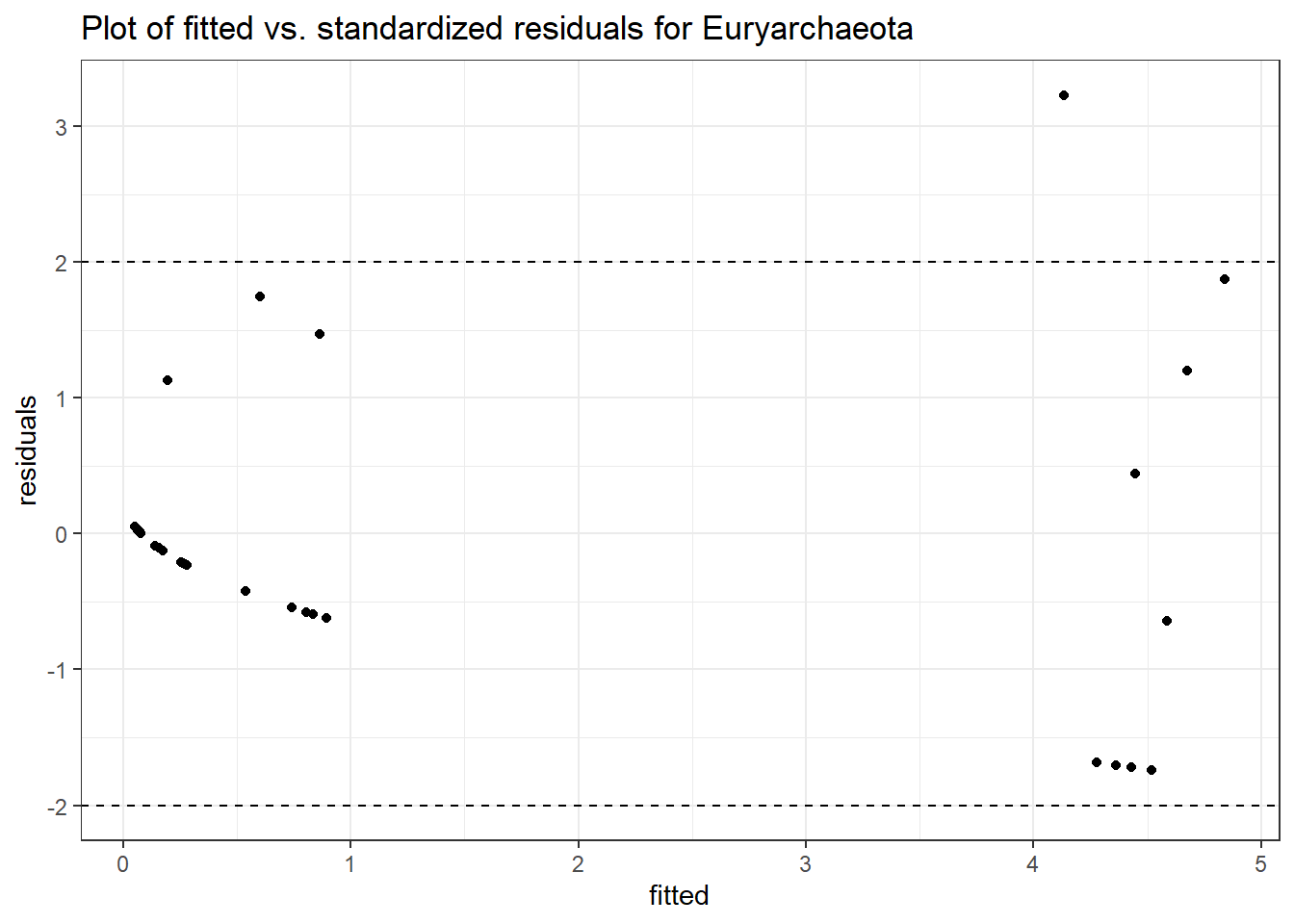

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -2.50184 1.11847 -2.23684 0.02530
intB Euryarchaeota 3.05713 1.24510 2.45533 0.01408
time Euryarchaeota 0.10681 0.41712 0.25607 0.79790
intB:time Euryarchaeota -0.14146 0.43994 -0.32153 0.74781
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.3002
Significance test not available for random effects
#########################################
#########################################
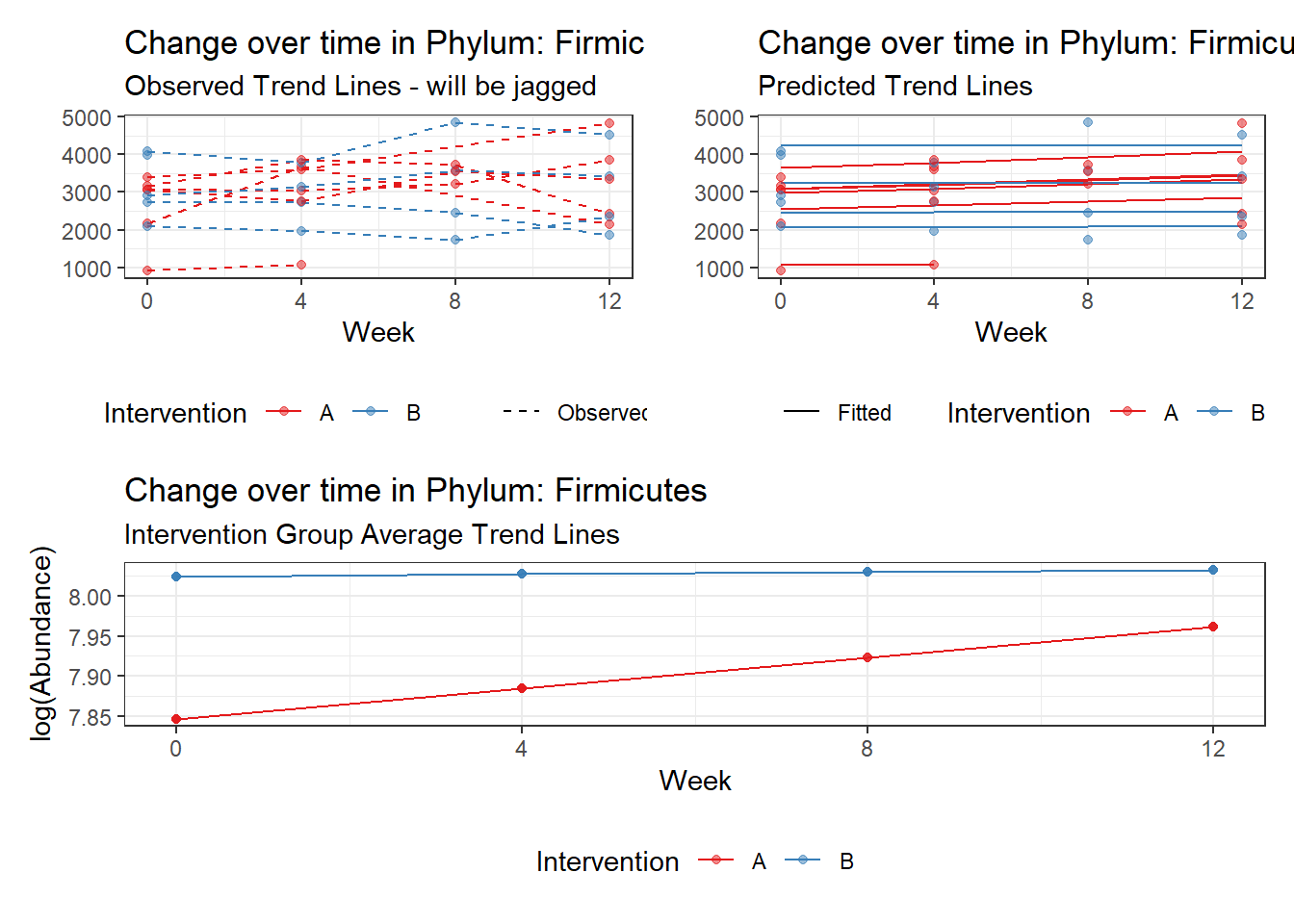
Model: Firmicutes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.1267112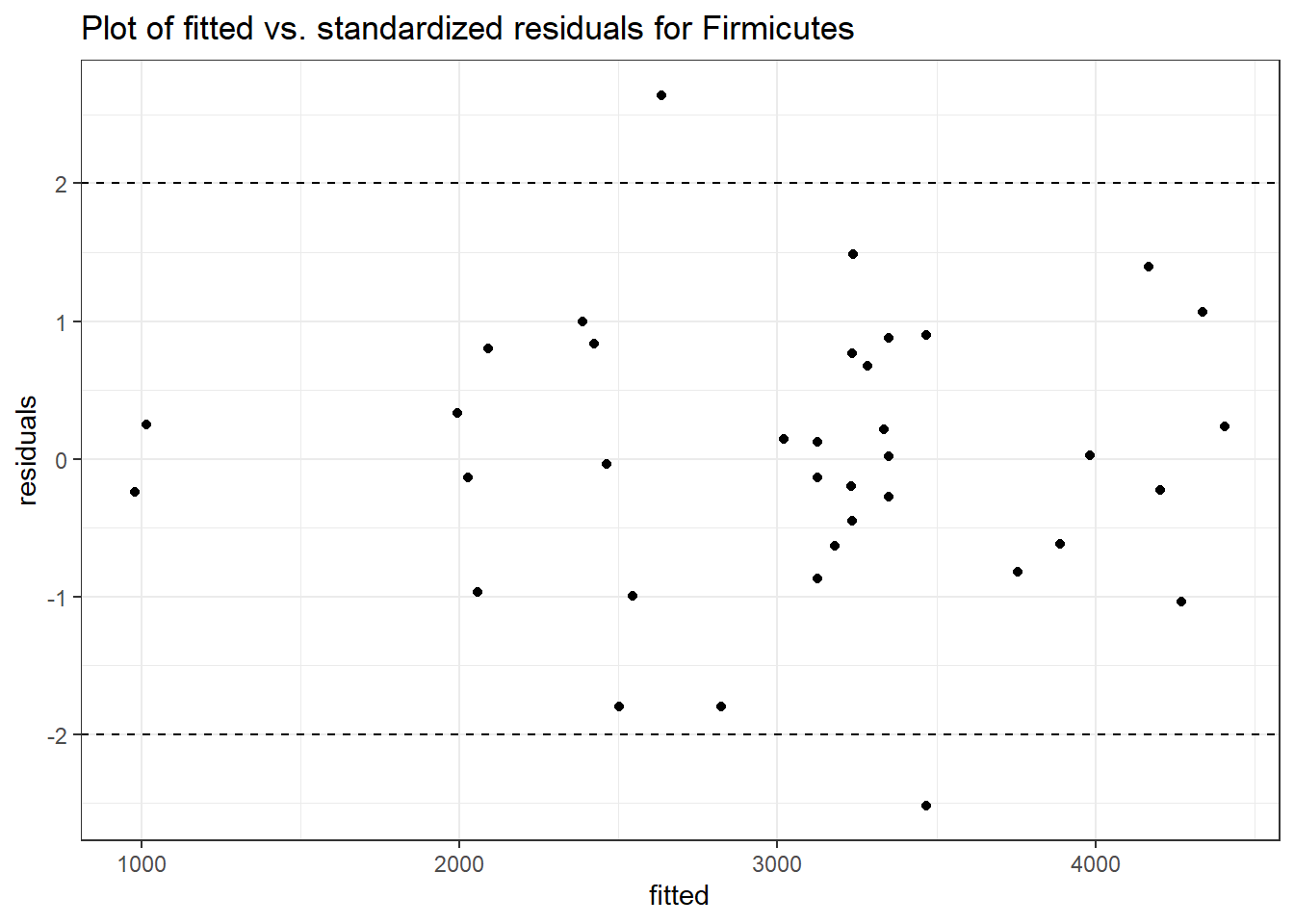

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.84451 0.15566 50.39470 0.00000
intB Firmicutes 0.17010 0.23087 0.73679 0.46125
time Firmicutes 0.03463 0.00352 9.83067 0.00000
intB:time Firmicutes -0.01883 0.00539 -3.49576 0.00047
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.38092
Significance test not available for random effects
#########################################
#########################################
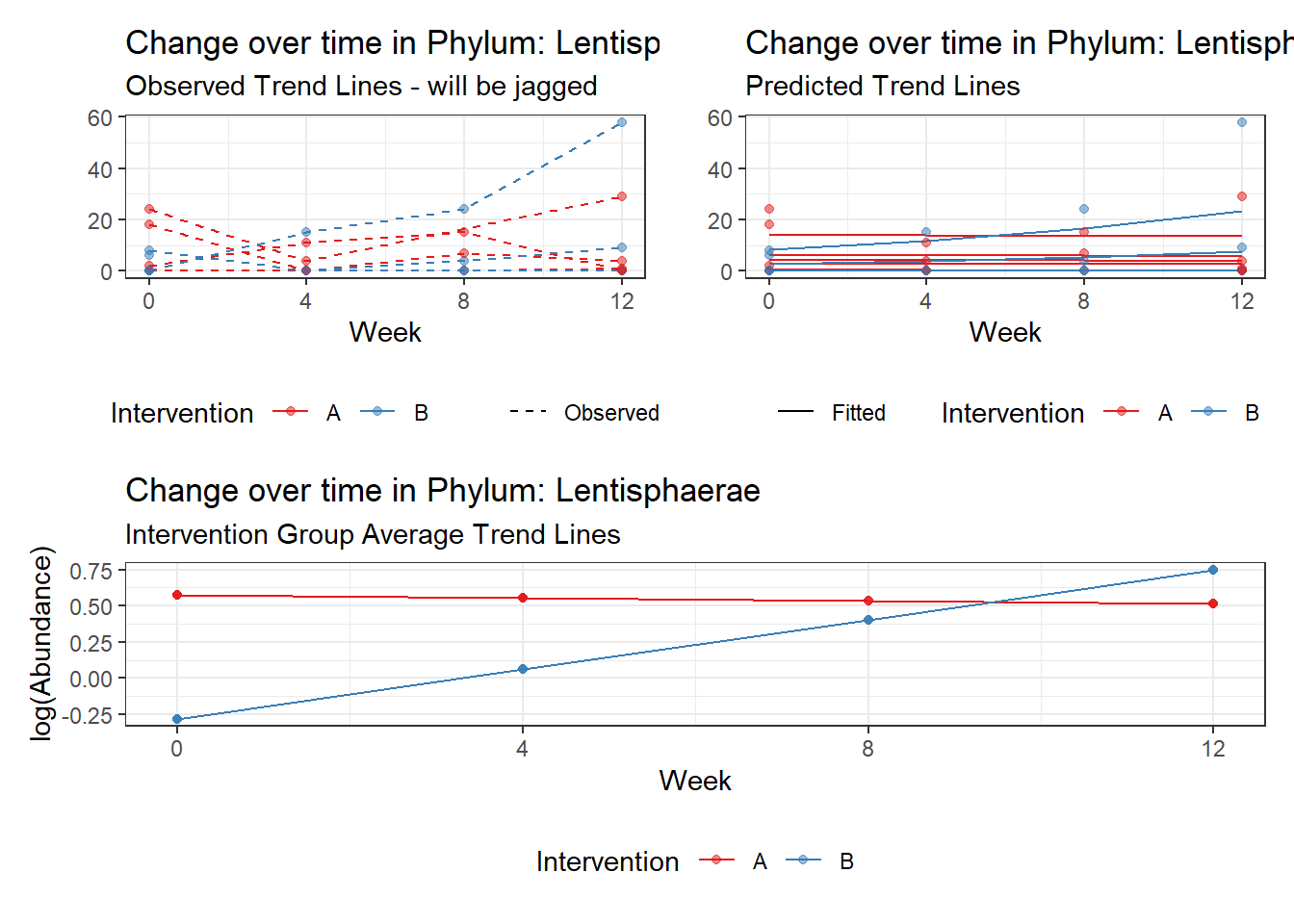
Model: Lentisphaerae ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.8722509

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.33280 1.19692 0.27805 0.78097
intB Lentisphaerae -1.36502 1.72220 -0.79260 0.42801
time Lentisphaerae -0.00972 0.07896 -0.12315 0.90199
intB:time Lentisphaerae 0.73924 0.12791 5.77923 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.613
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.07355022
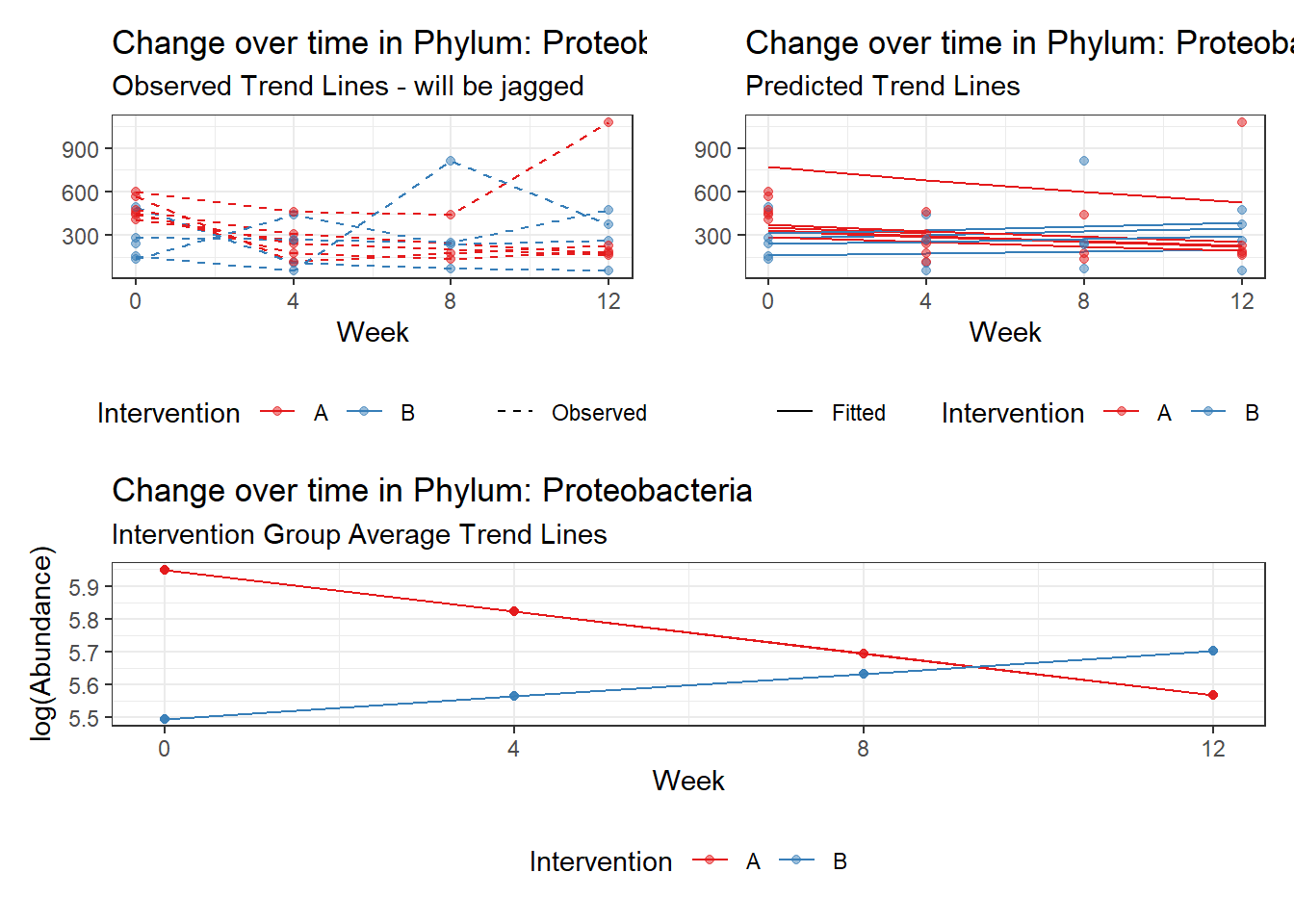
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.94829 0.11641 51.09812 0.00000
intB Proteobacteria -0.45370 0.17331 -2.61791 0.00885
time Proteobacteria -0.12701 0.01087 -11.68390 0.00000
intB:time Proteobacteria 0.19592 0.01725 11.35999 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.28176
Significance test not available for random effects
#########################################
#########################################
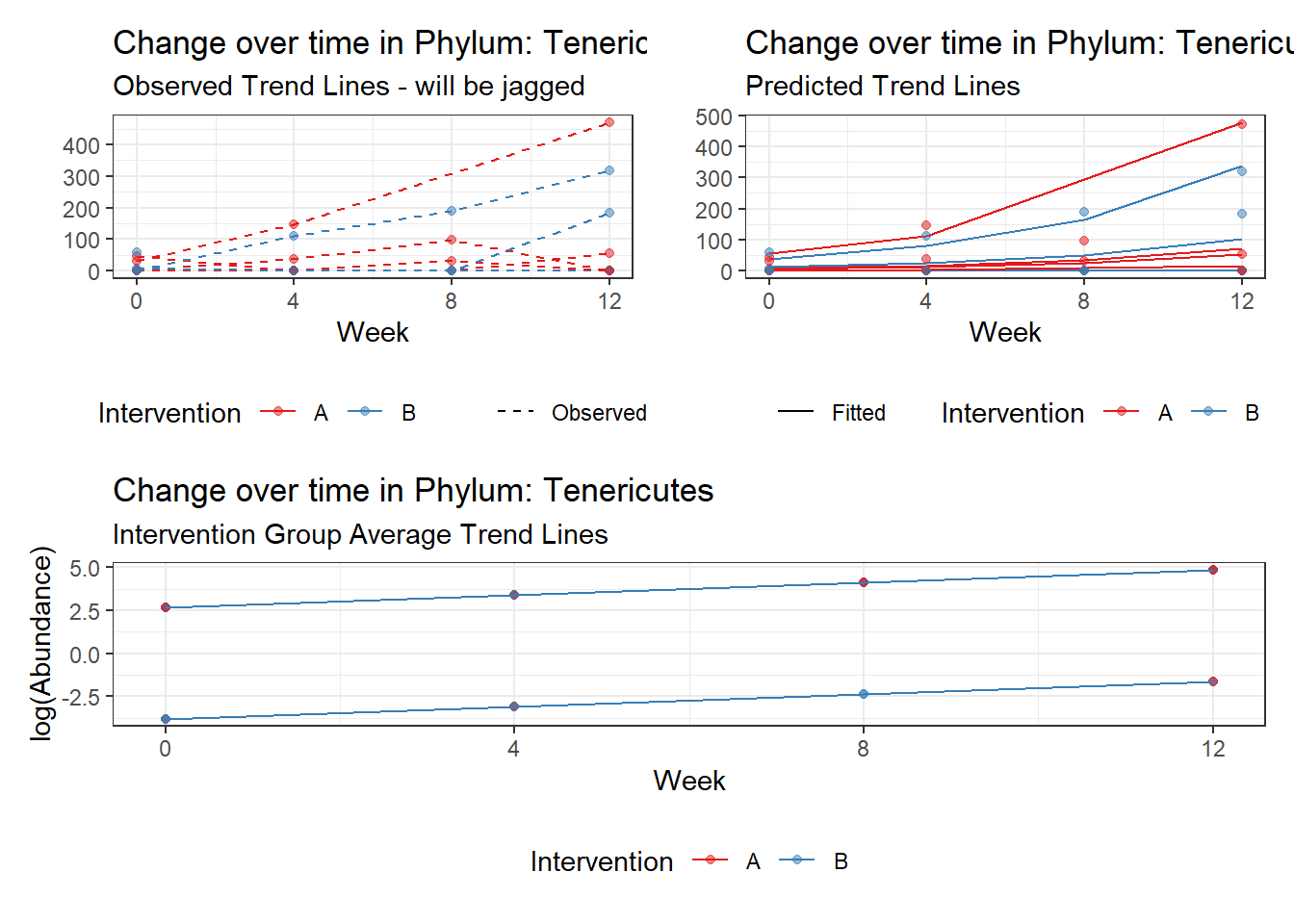
Model: Tenericutes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.9207144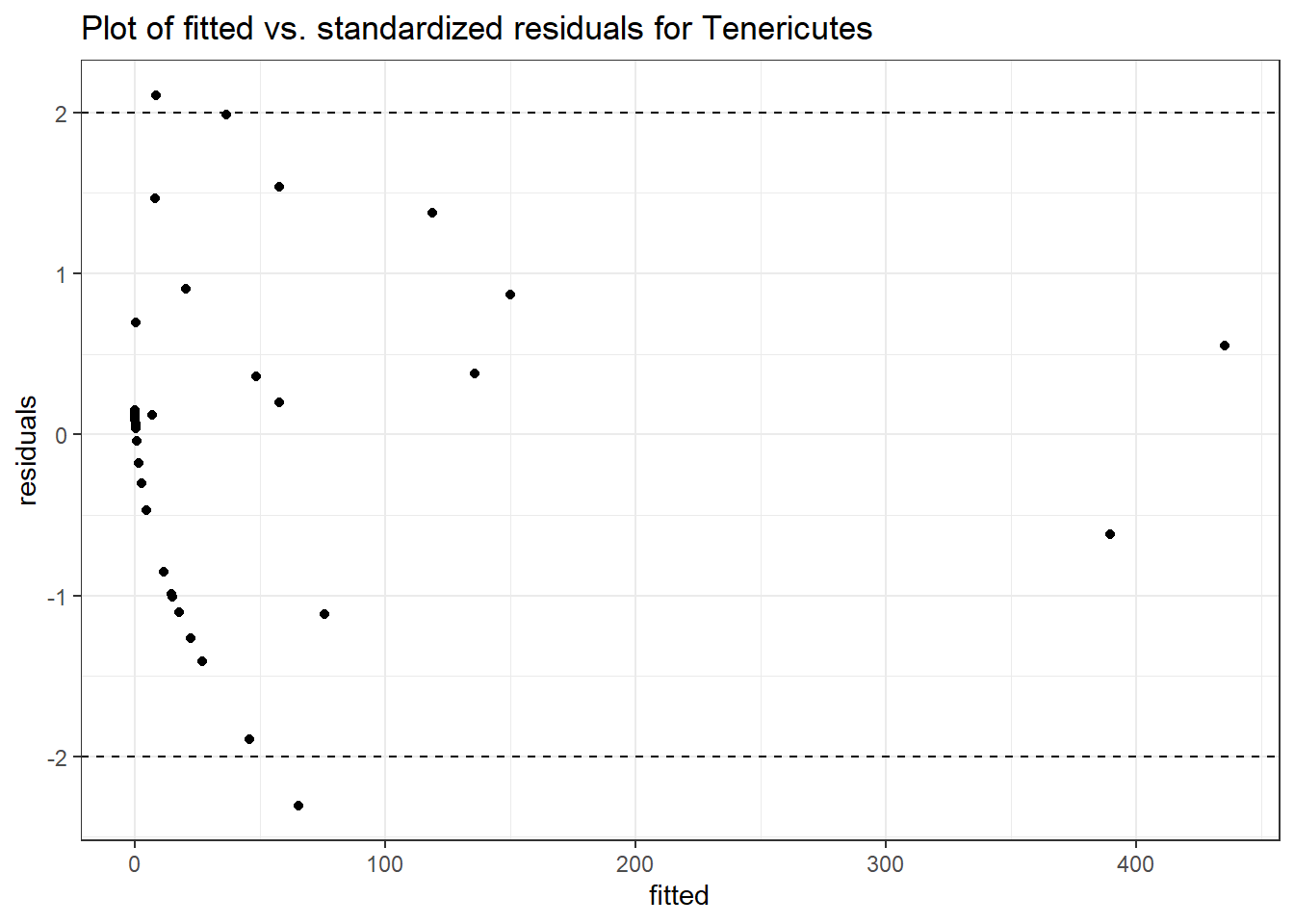
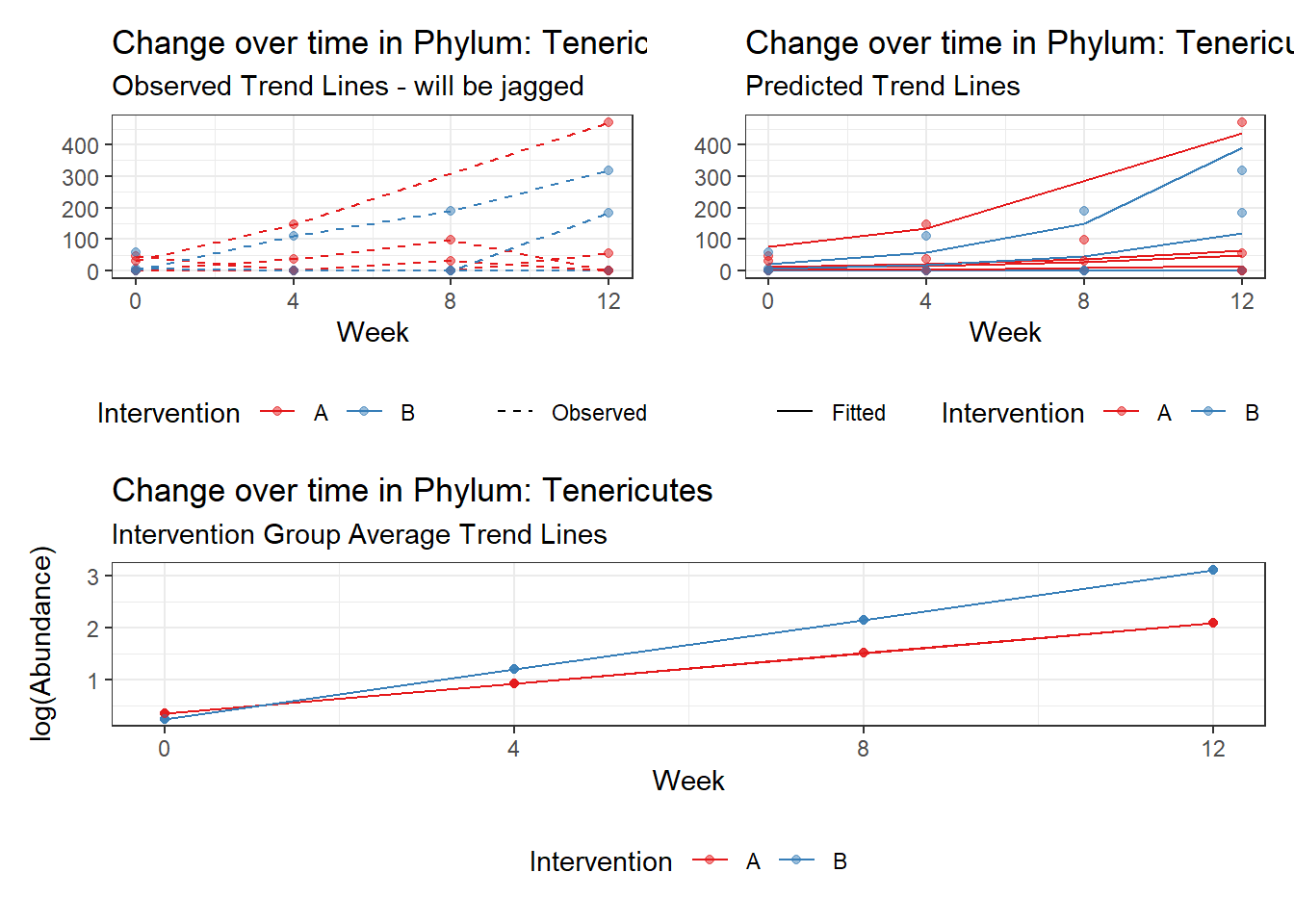
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 0.34347 1.45013 0.23685 0.81277
intB Tenericutes -0.09897 2.12036 -0.04668 0.96277
time Tenericutes 0.58336 0.03024 19.28914 0.00000
intB:time Tenericutes 0.37119 0.05296 7.00871 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.4077
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x0000000043de42d0>
Link: poisson
Intraclass Correlation (ICC): 0.8554283
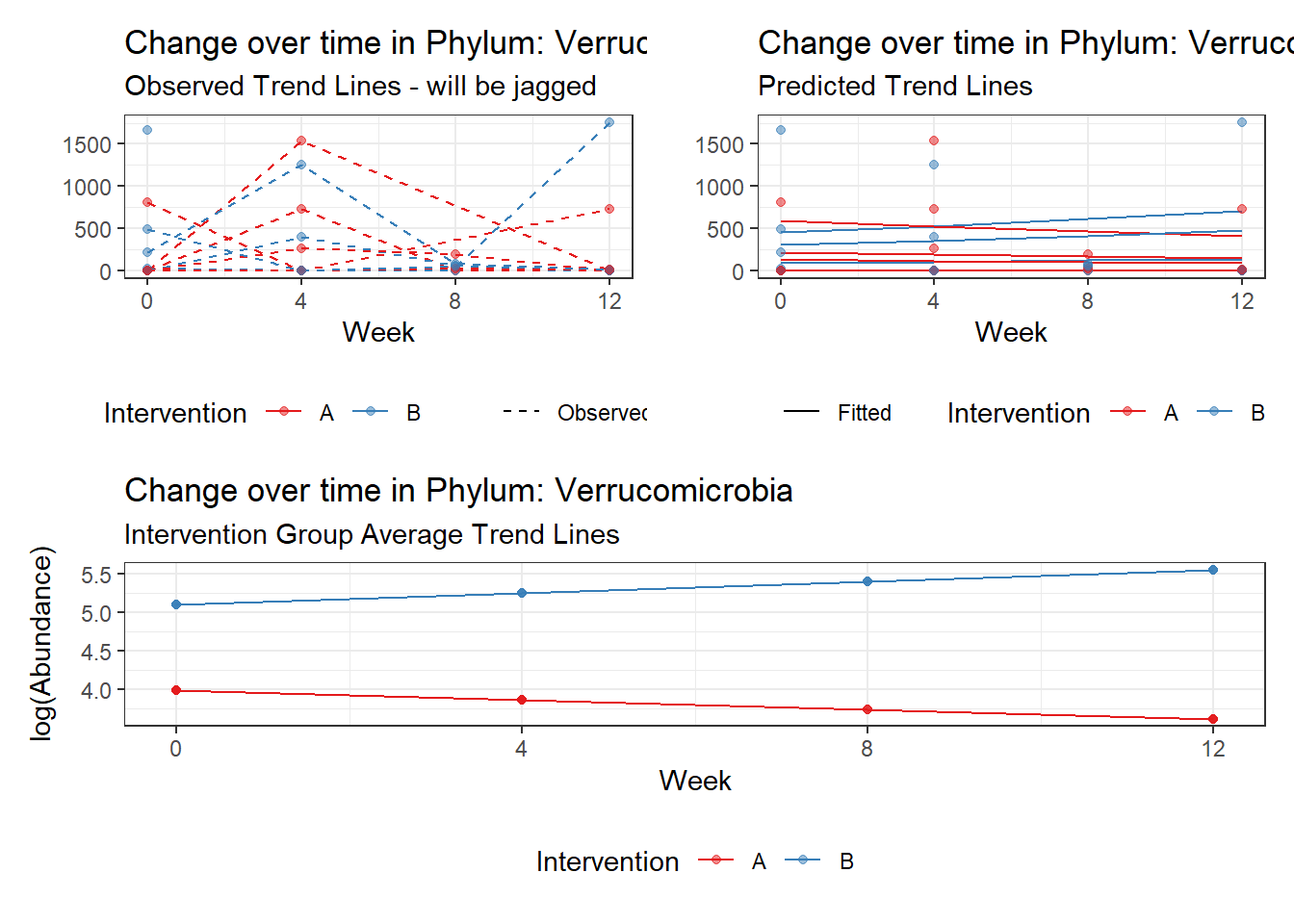
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 3.98707 1.00988 3.94807 0.00008
intB Verrucomicrobia 1.11249 1.48508 0.74911 0.45379
time Verrucomicrobia -0.12308 0.01296 -9.49425 0.00000
intB:time Verrucomicrobia 0.27329 0.01883 14.51628 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4325
Significance test not available for random effectsphylum.fit5b <- glmm_microbiome(mydata=microbiome_data,model.number=5,
taxa.level="Phylum", link="negbinom",
model="1 + intB + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinom
Intraclass Correlation (ICC): 0.3032304
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.19349 0.51139 4.28922 0.00002
intB Actinobacteria 0.69673 0.75295 0.92533 0.35480
time Actinobacteria 0.13647 0.24738 0.55168 0.58117
intB:time Actinobacteria 0.12973 0.37276 0.34802 0.72782
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.65969
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinom
Intraclass Correlation (ICC): 0.1379942
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.14348 0.18180 44.79293 0.00000
intB Bacteroidetes -0.21784 0.27192 -0.80113 0.42305
time Bacteroidetes -0.04041 0.04609 -0.87681 0.38059
intB:time Bacteroidetes -0.01395 0.07007 -0.19911 0.84218
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.40011
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced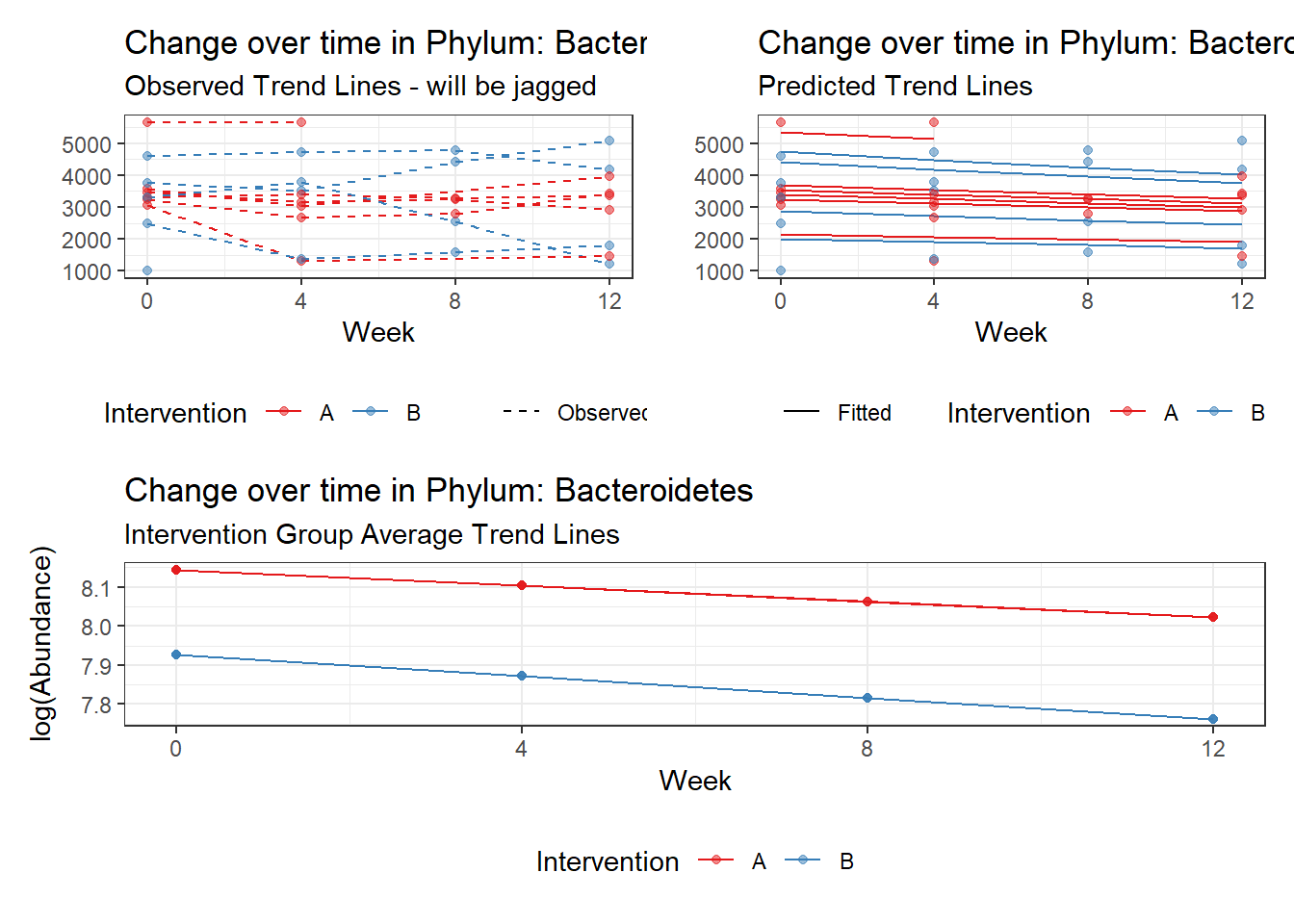
Intraclass Correlation (ICC): 0.9443394
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -1.69265 2.28381 -0.74115 0.45860
intB Cyanobacteria 1.85125 2.85158 0.64920 0.51621
time Cyanobacteria -0.07222 0.04213 -1.71399 0.08653
intB:time Cyanobacteria -0.50485 0.06903 -7.31379 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.119
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinomboundary (singular) fit: see ?isSingular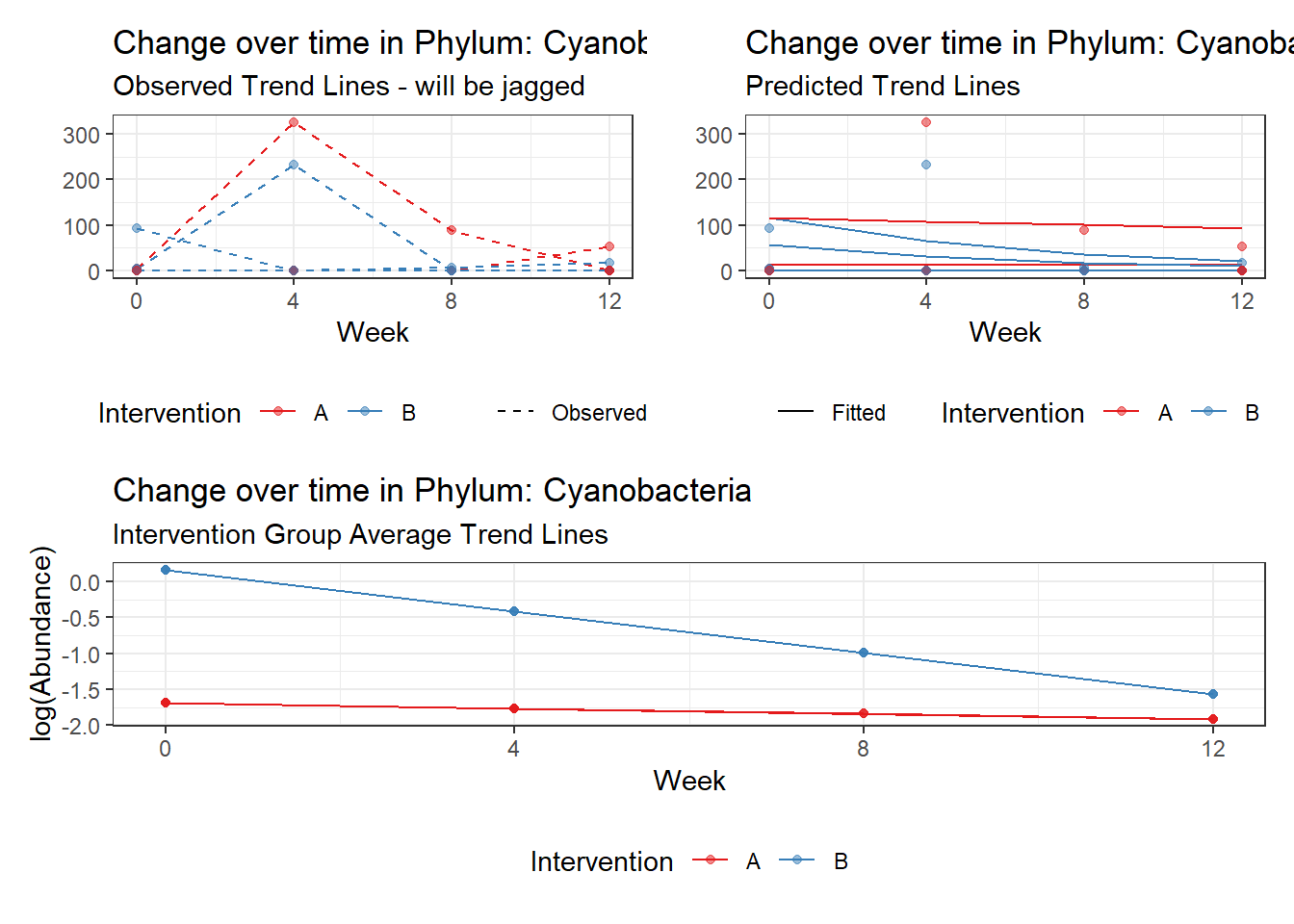
Intraclass Correlation (ICC): 0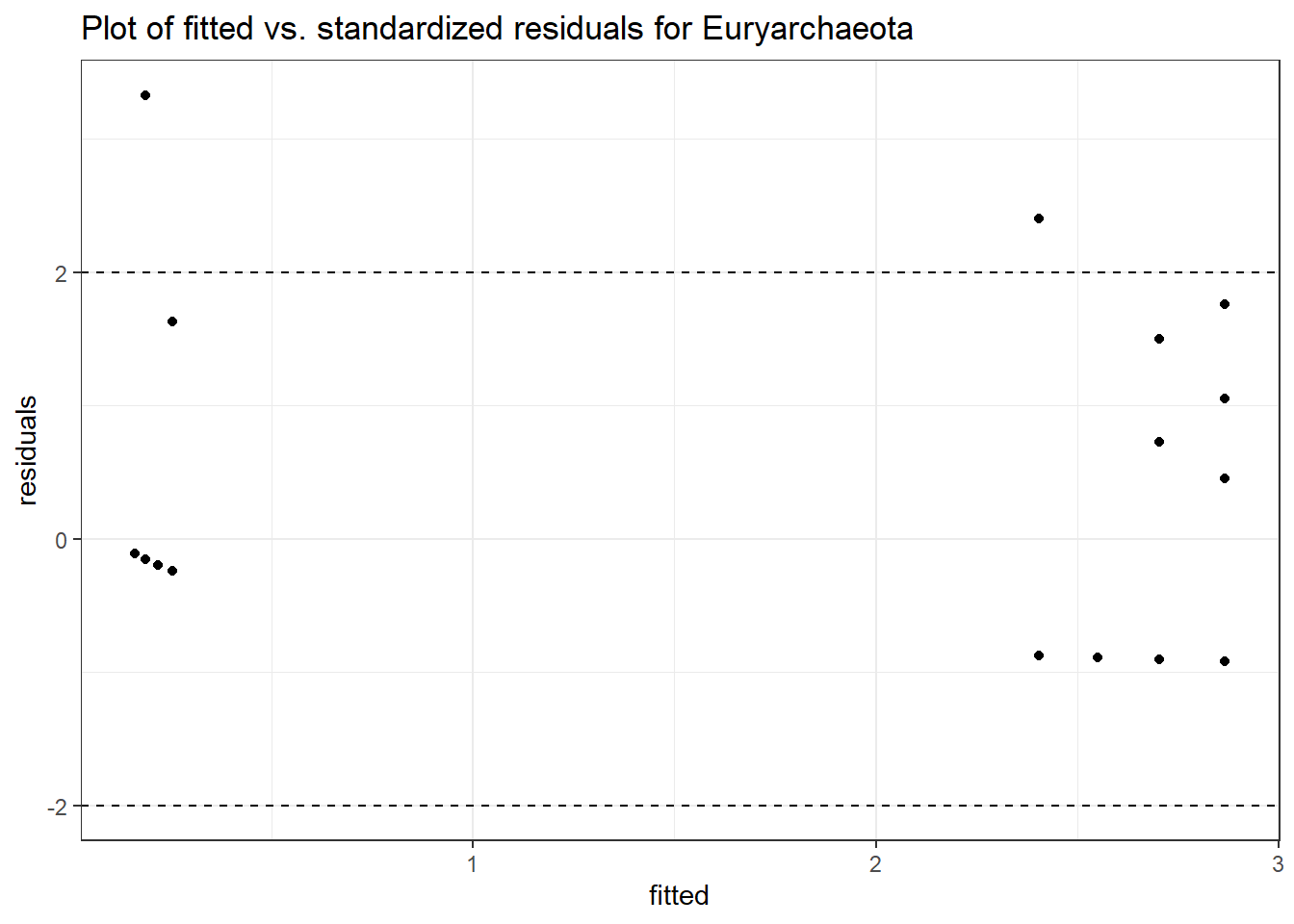
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -1.82395 1.32482 -1.37675 0.16859
intB Euryarchaeota 2.87704 1.62156 1.77424 0.07602
time Euryarchaeota 0.15064 0.76730 0.19633 0.84435
intB:time Euryarchaeota -0.20915 0.90786 -0.23038 0.81780
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinom
Intraclass Correlation (ICC): 0.115014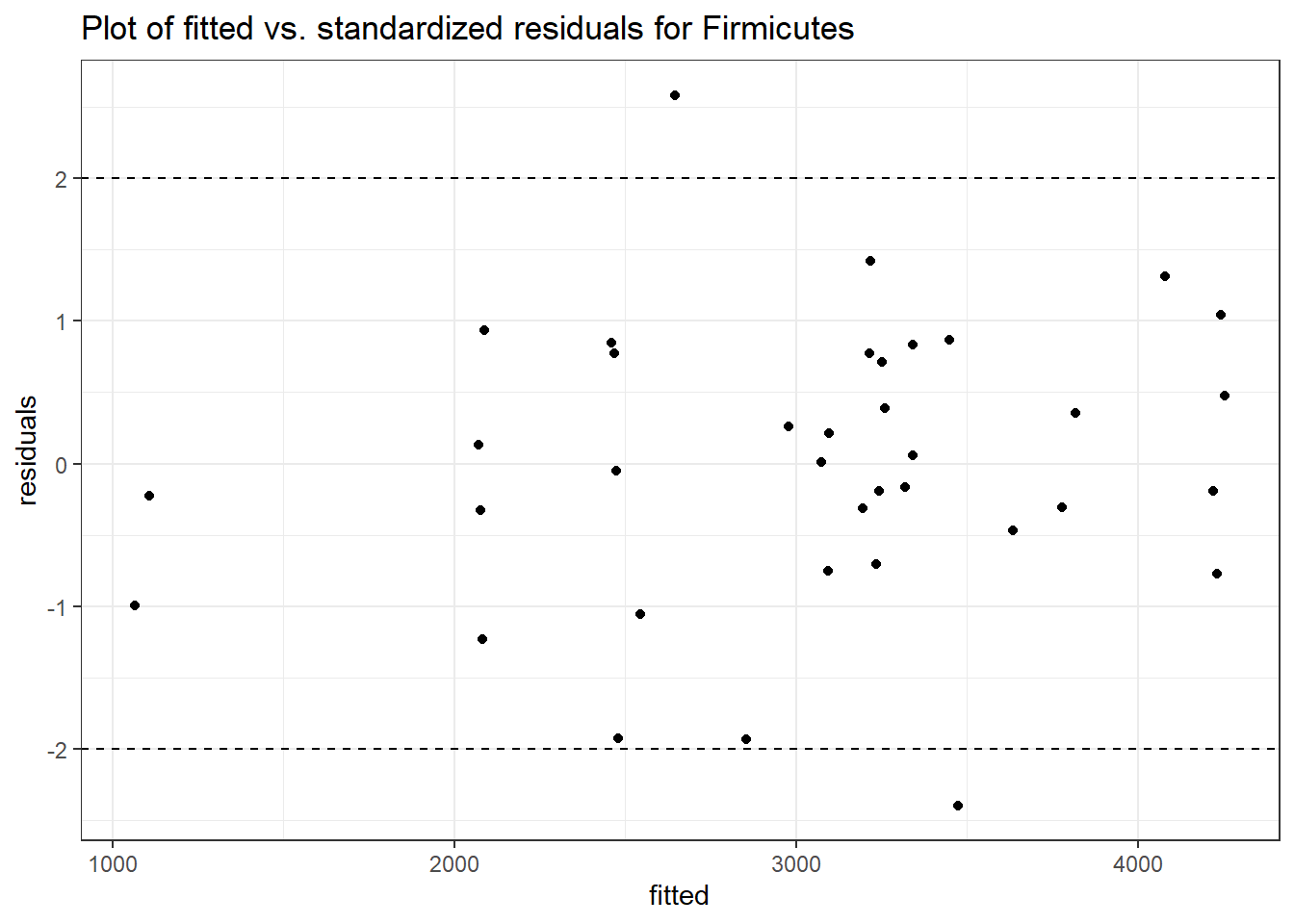
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.84659 0.15736 49.86502 0.00000
intB Firmicutes 0.17778 0.23362 0.76098 0.44667
time Firmicutes 0.03833 0.03259 1.17590 0.23963
intB:time Firmicutes -0.03570 0.04825 -0.73979 0.45942
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.3605
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinom
Intraclass Correlation (ICC): 0.8191818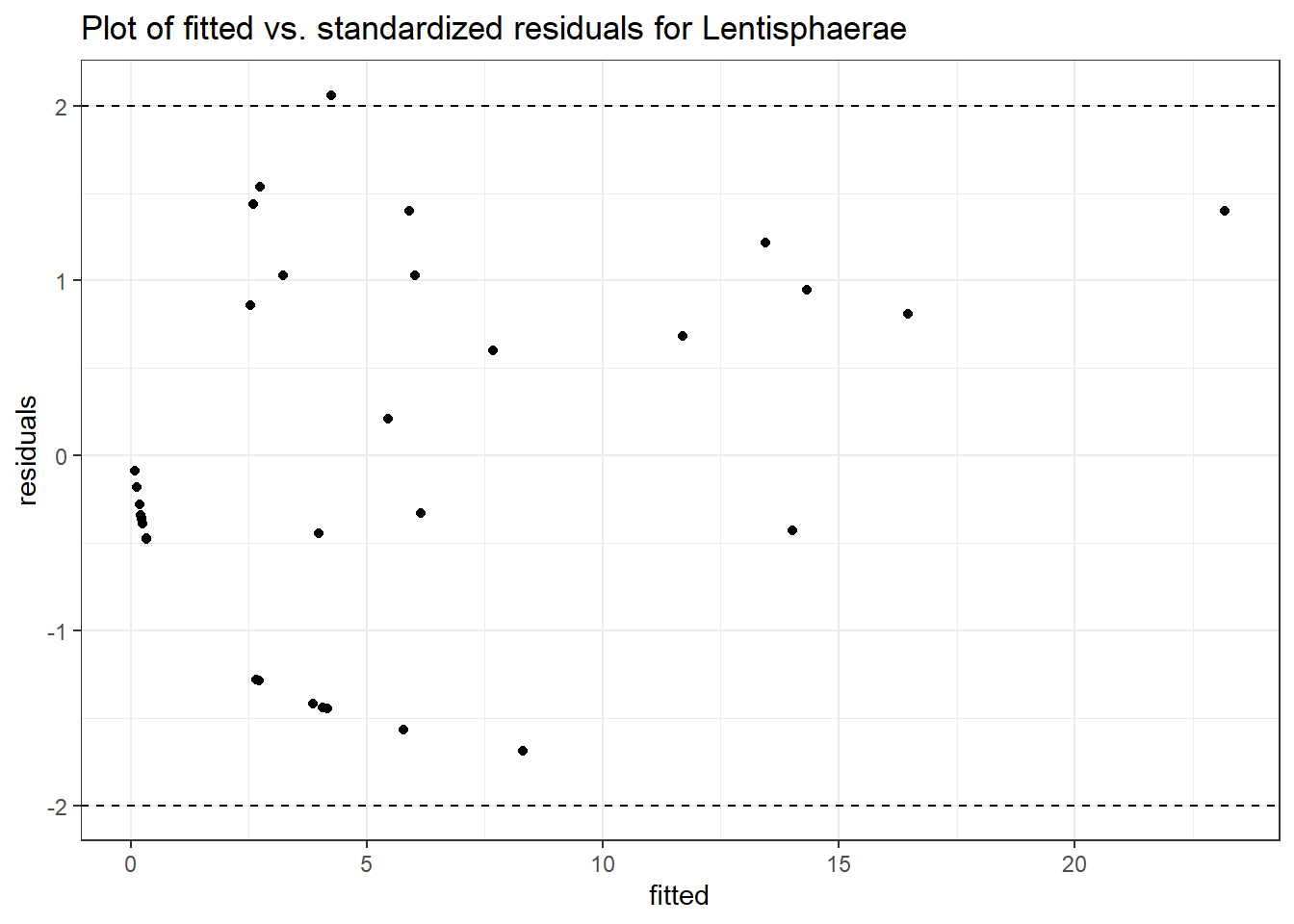
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.57683 1.17881 0.48933 0.62461
intB Lentisphaerae -0.85736 1.67372 -0.51225 0.60848
time Lentisphaerae -0.02082 0.37741 -0.05515 0.95602
intB:time Lentisphaerae 0.36326 0.55644 0.65282 0.51387
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1285
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinom
Intraclass Correlation (ICC): 0.02837805
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.98502 0.20762 28.82736 0.00000
intB Proteobacteria -0.45039 0.32063 -1.40473 0.16010
time Proteobacteria -0.12373 0.11430 -1.08256 0.27901
intB:time Proteobacteria 0.17719 0.16924 1.04695 0.29512
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.1709
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinomWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.00438541 (tol = 0.002, component 1)boundary (singular) fit: see ?isSingular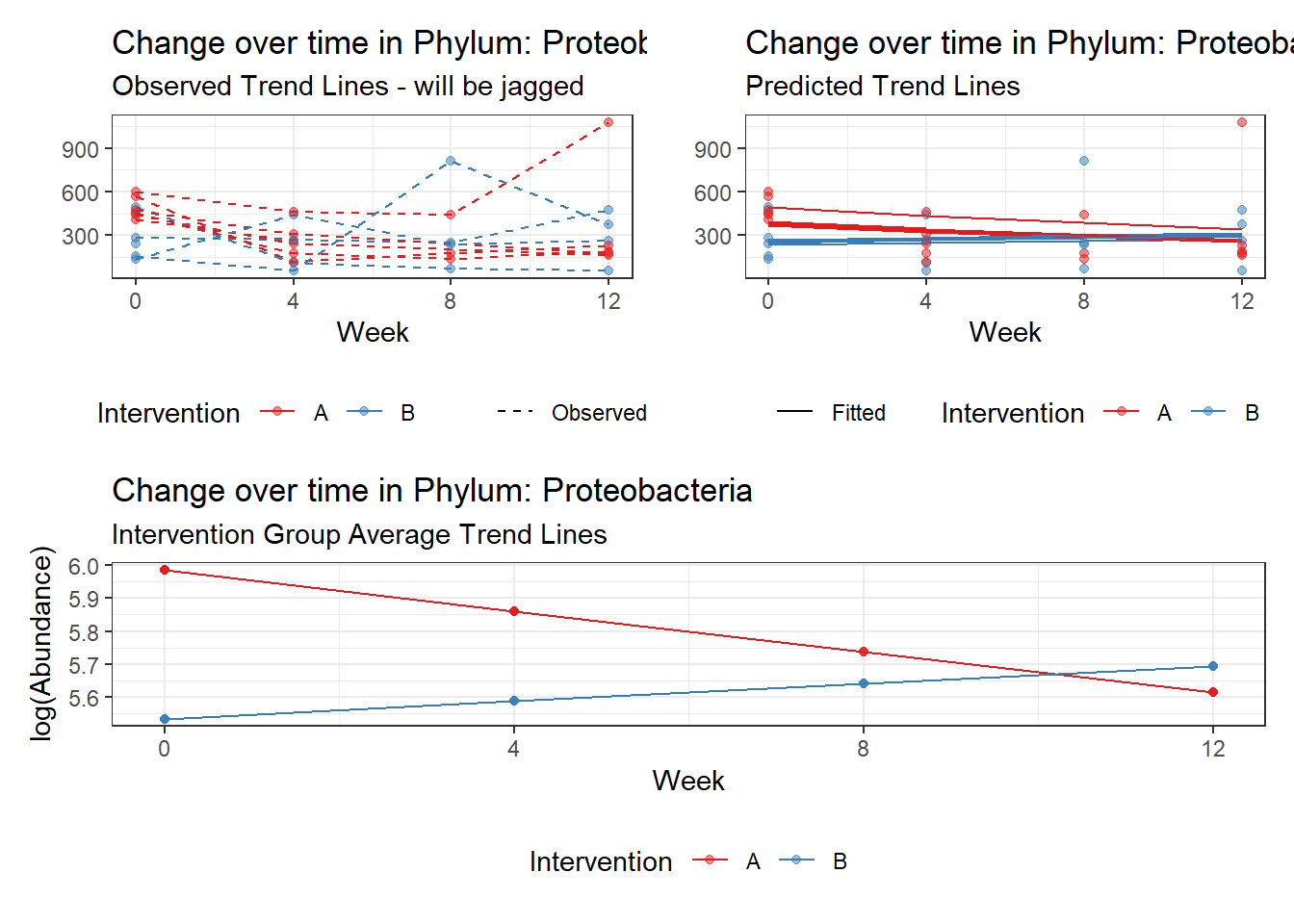
Intraclass Correlation (ICC): 4.481095e-12
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 2.59705 1.09668 2.36811 0.01788
intB Tenericutes -0.05400 1.63231 -0.03308 0.97361
time Tenericutes 0.68027 0.62041 1.09649 0.27286
intB:time Tenericutes 0.05798 0.90675 0.06394 0.94902
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1169e-06
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x000000004aef09d0>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 1.976585e-11

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 5.56968 0.96093 5.79616 0.00000
intB Verrucomicrobia 0.45954 1.33004 0.34551 0.72971
time Verrucomicrobia -0.15489 0.57324 -0.27020 0.78701
intB:time Verrucomicrobia 0.03610 0.74911 0.04819 0.96157
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.4459e-06
Significance test not available for random effectsCovariate(s): Time by intervention interaction (only)
Only the interaction term.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
phylum.fit6 <- glmm_microbiome(mydata=microbiome_data,model.number=6,
taxa.level="Phylum", link="poisson",
model="1 + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.4783296
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.53592 0.29696 8.53961 0.00000
time Actinobacteria 0.09052 0.04830 1.87418 0.06091
time:intB Actinobacteria 0.14692 0.06211 2.36544 0.01801
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.95756
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.1811897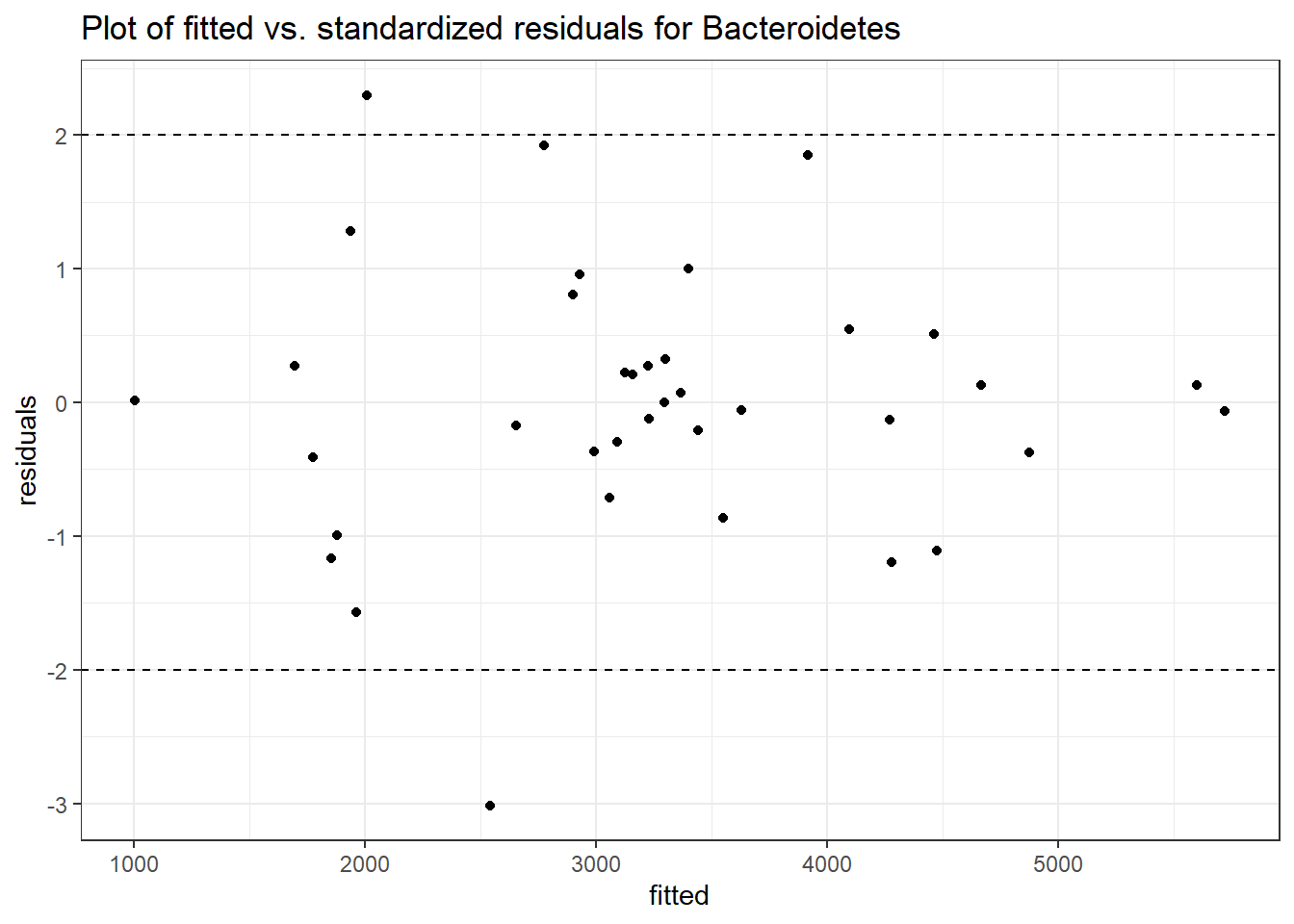
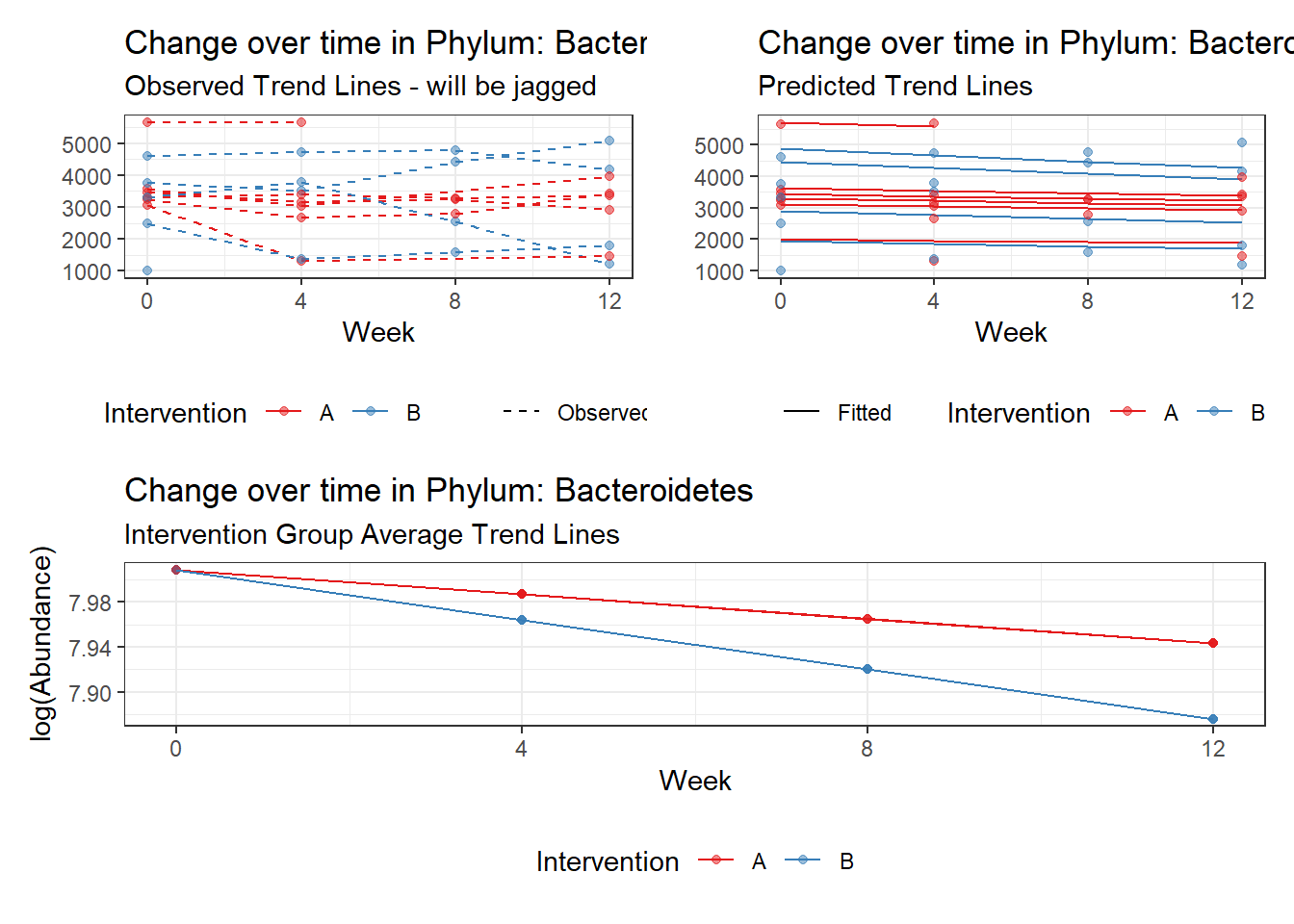
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.00875 0.14193 56.42900 0e+00
time Bacteroidetes -0.02181 0.00363 -6.00767 0e+00
time:intB Bacteroidetes -0.02248 0.00531 -4.22991 2e-05
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.47041
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.9459588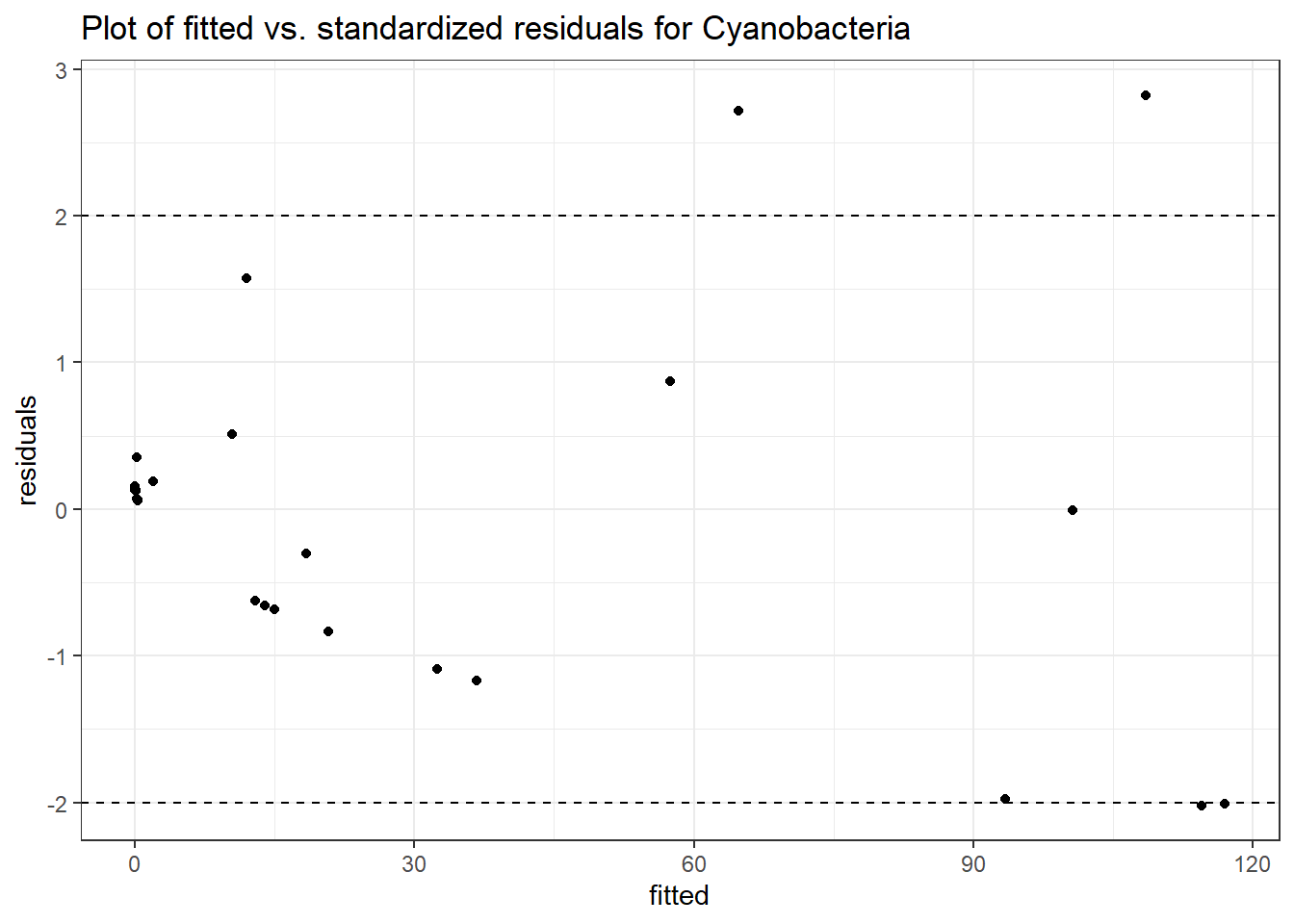
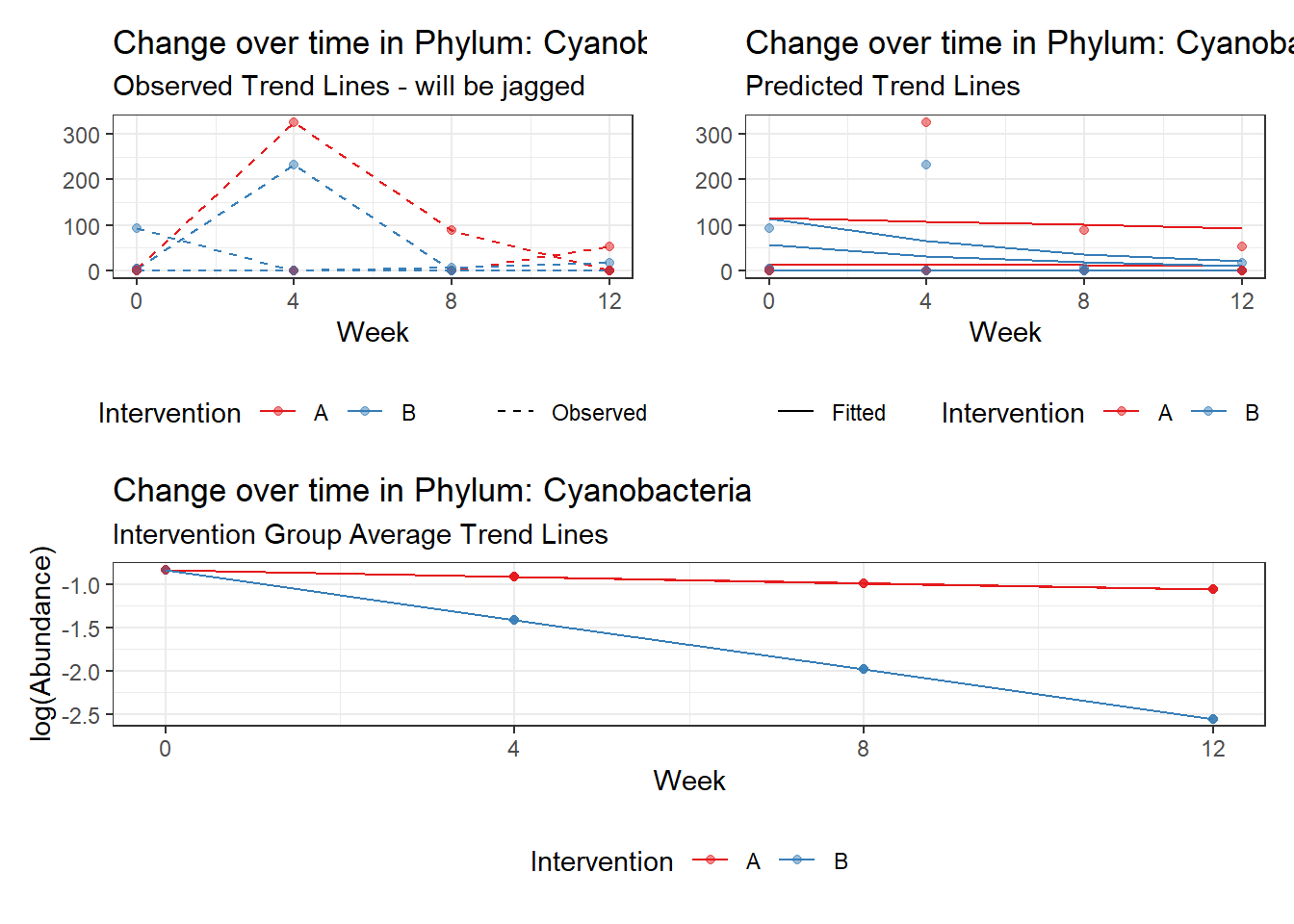
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -0.83959 1.71059 -0.49082 0.62355
time Cyanobacteria -0.07509 0.04112 -1.82584 0.06787
time:intB Cyanobacteria -0.49494 0.06783 -7.29656 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.1838
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.7947804
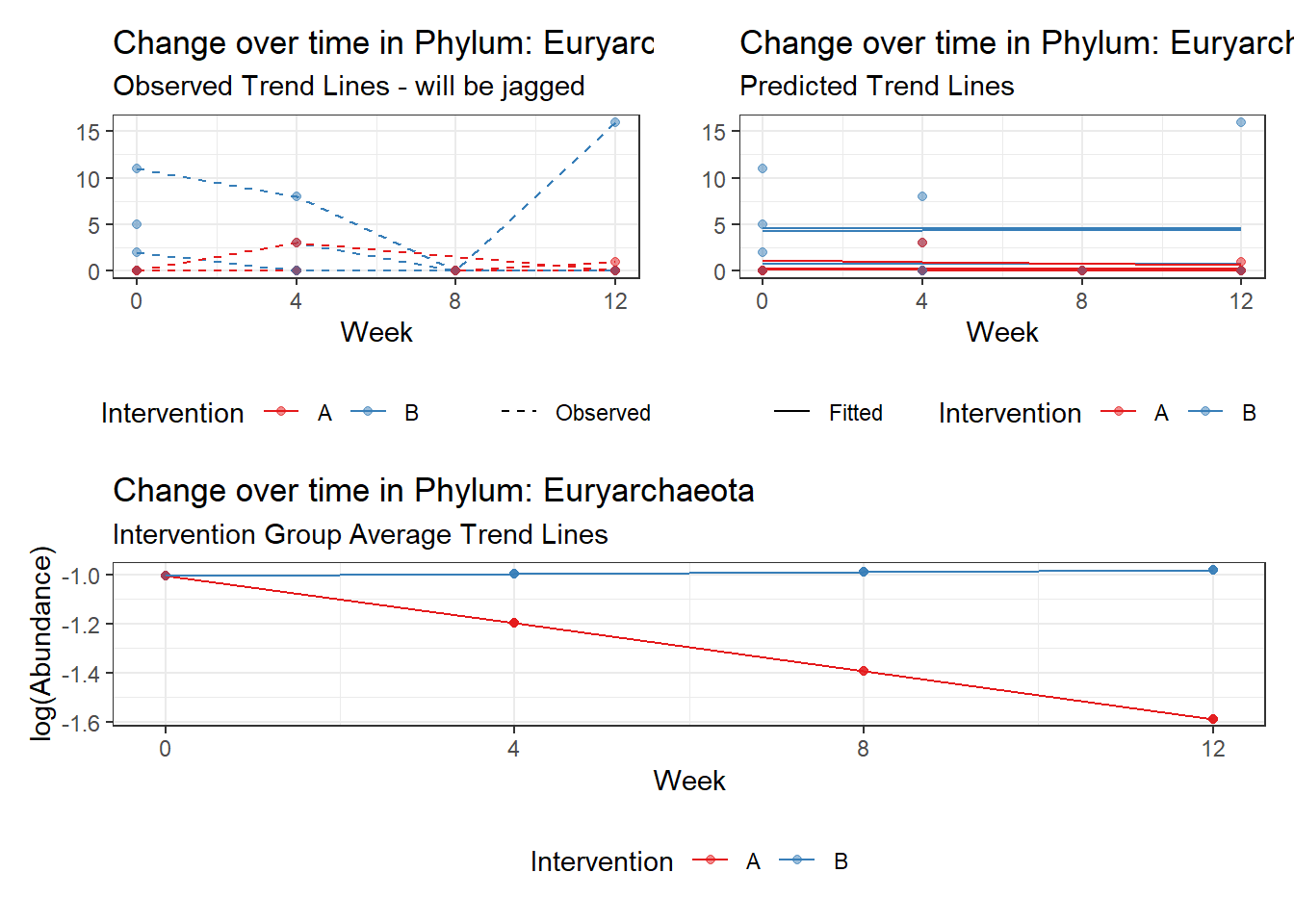
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -1.00544 0.88489 -1.13623 0.25586
time Euryarchaeota -0.19368 0.38486 -0.50324 0.61480
time:intB Euryarchaeota 0.20135 0.40971 0.49143 0.62312
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.968
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.1321344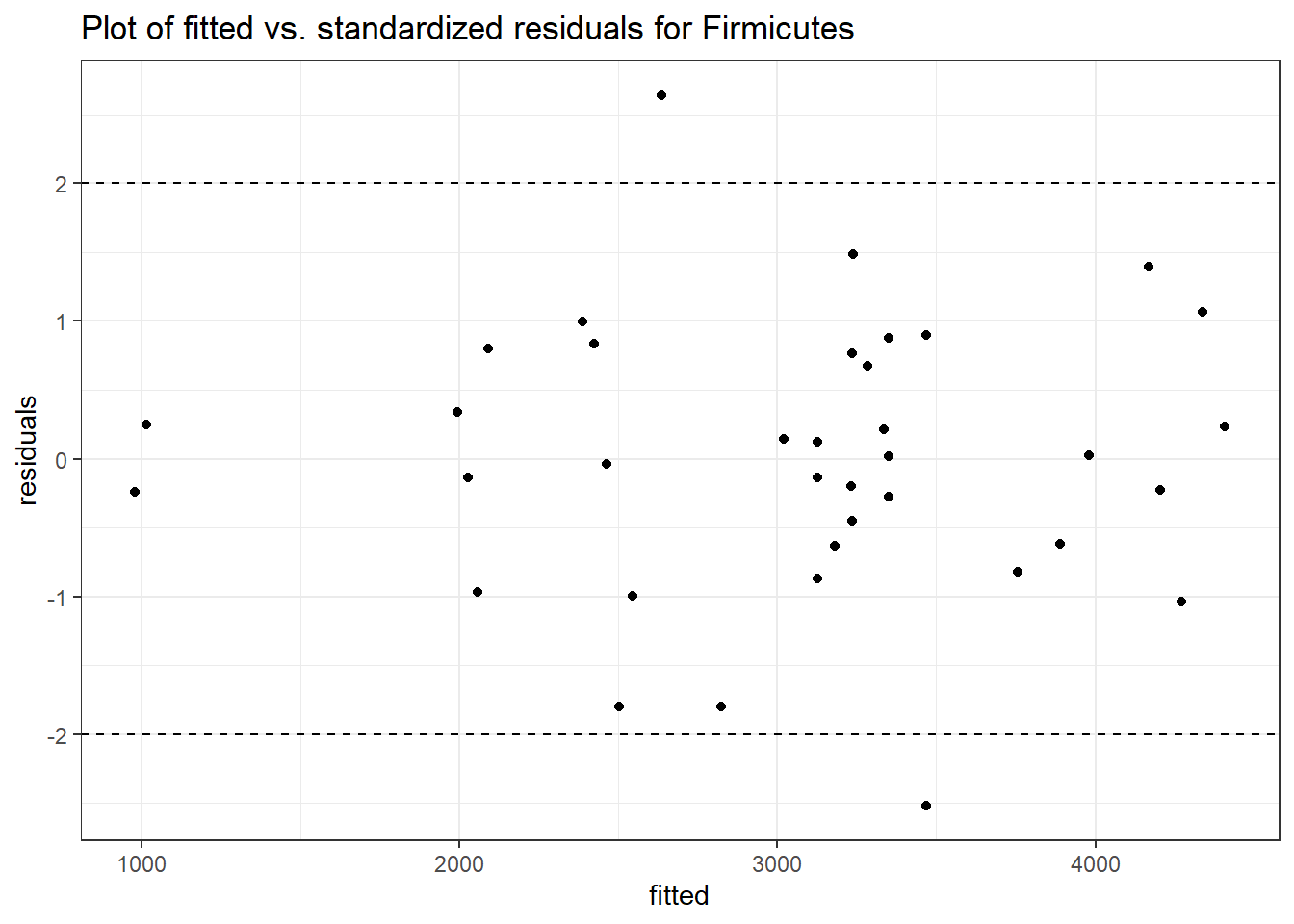
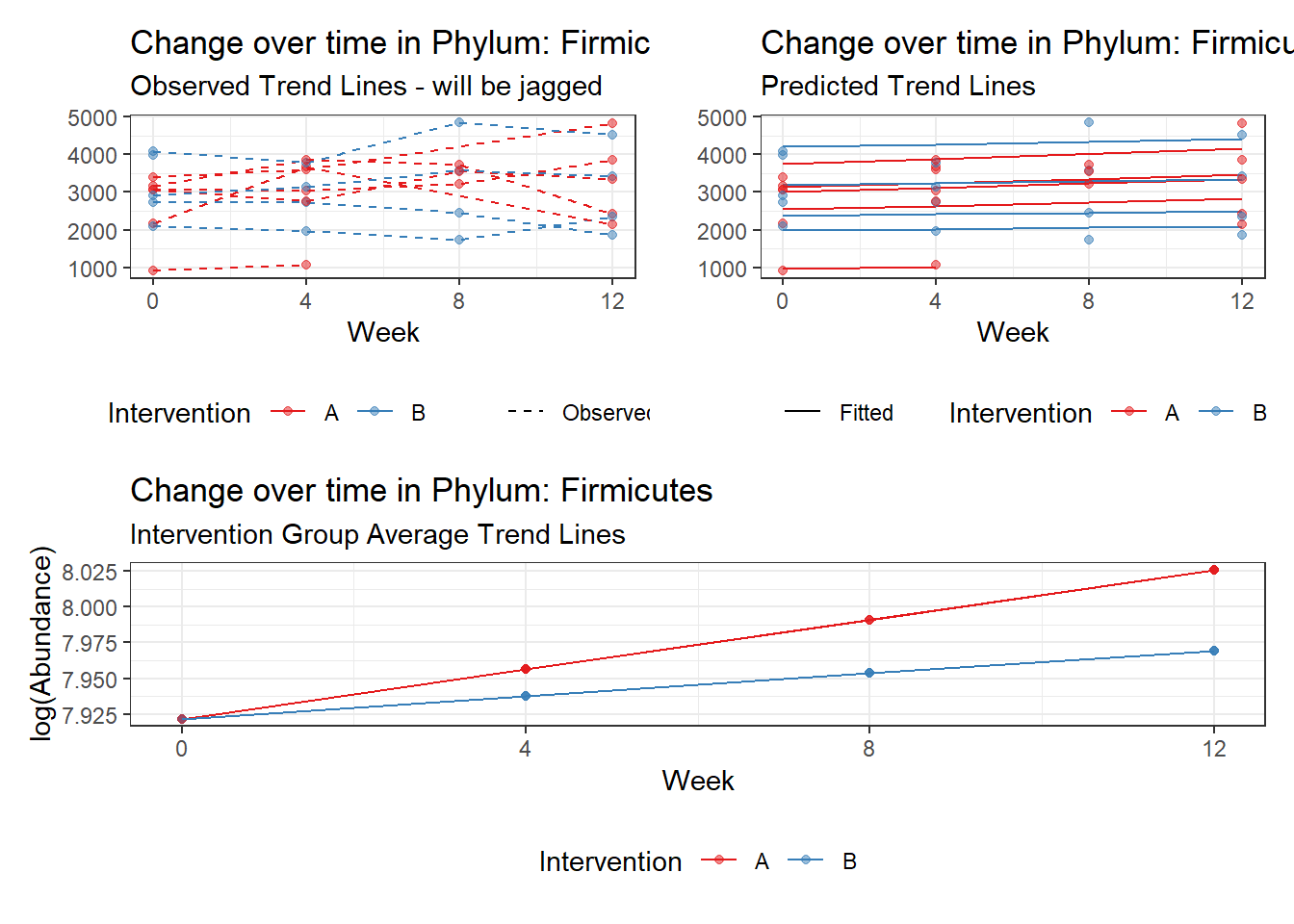
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.92184 0.11775 67.27597 0.00000
time Firmicutes 0.03457 0.00352 9.81800 0.00000
time:intB Firmicutes -0.01871 0.00538 -3.47559 0.00051
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.39019
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.8836954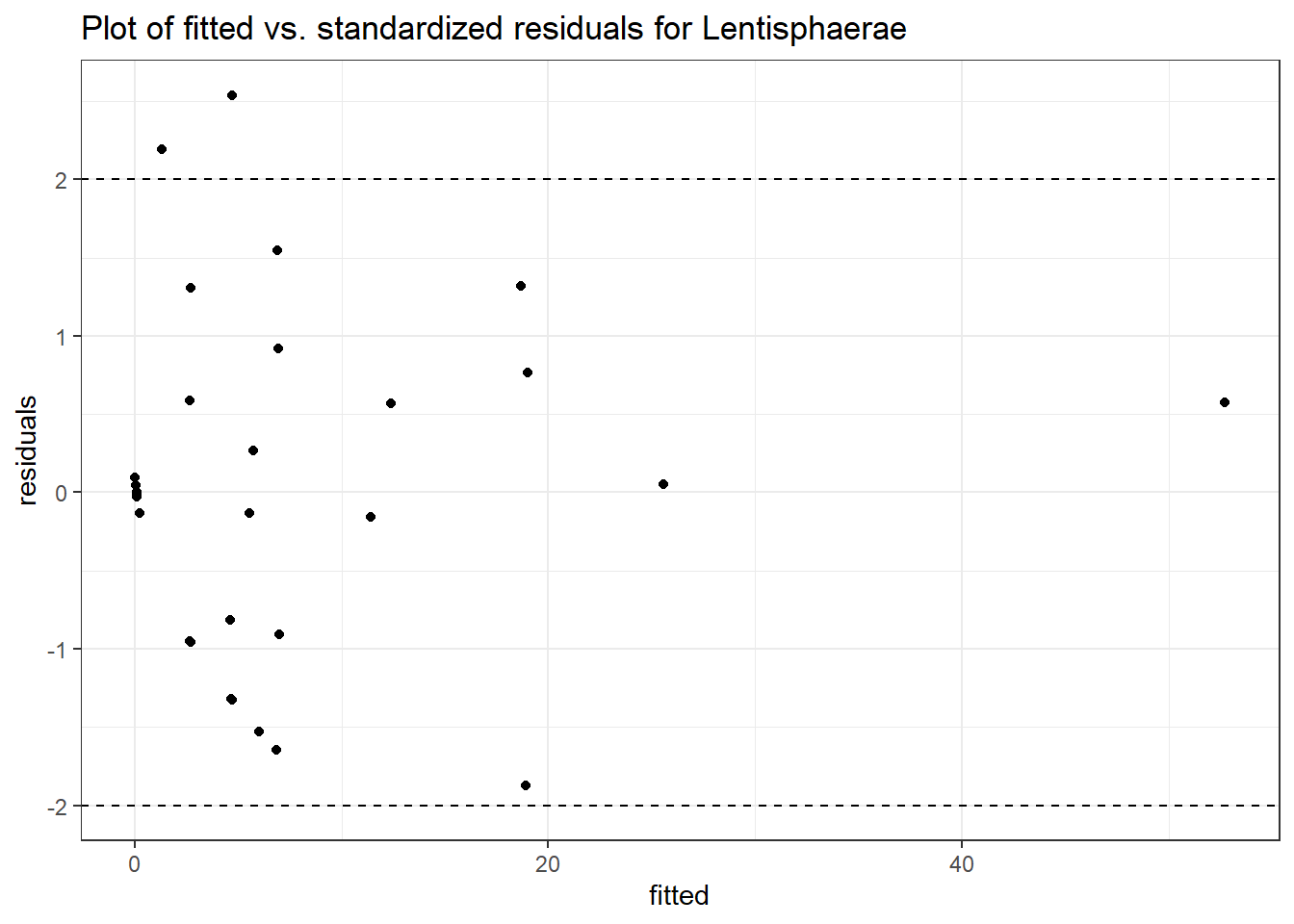
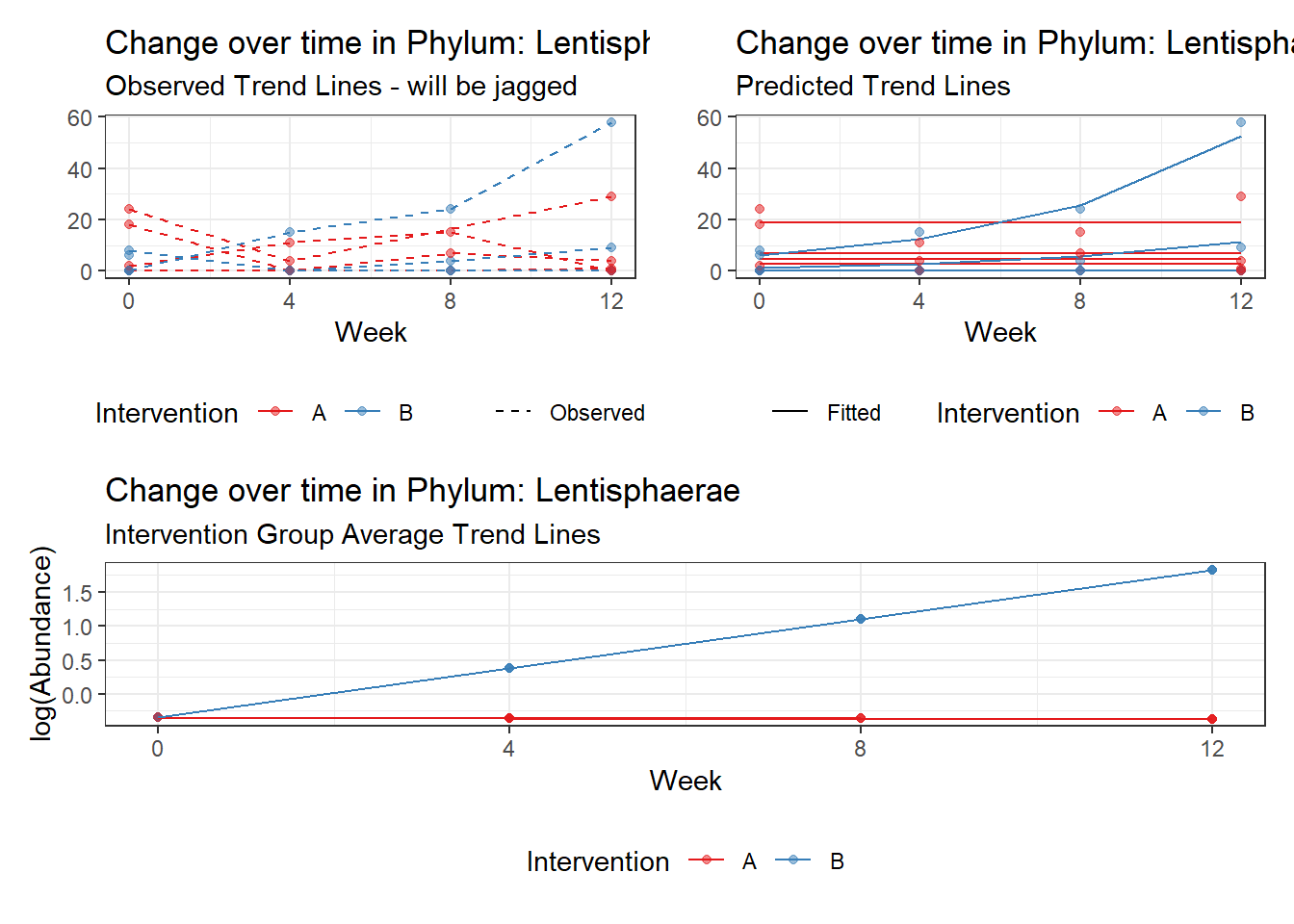
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae -0.34507 0.97811 -0.35279 0.72425
time Lentisphaerae -0.00644 0.07897 -0.08150 0.93504
time:intB Lentisphaerae 0.72920 0.12722 5.73197 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.7565
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.114292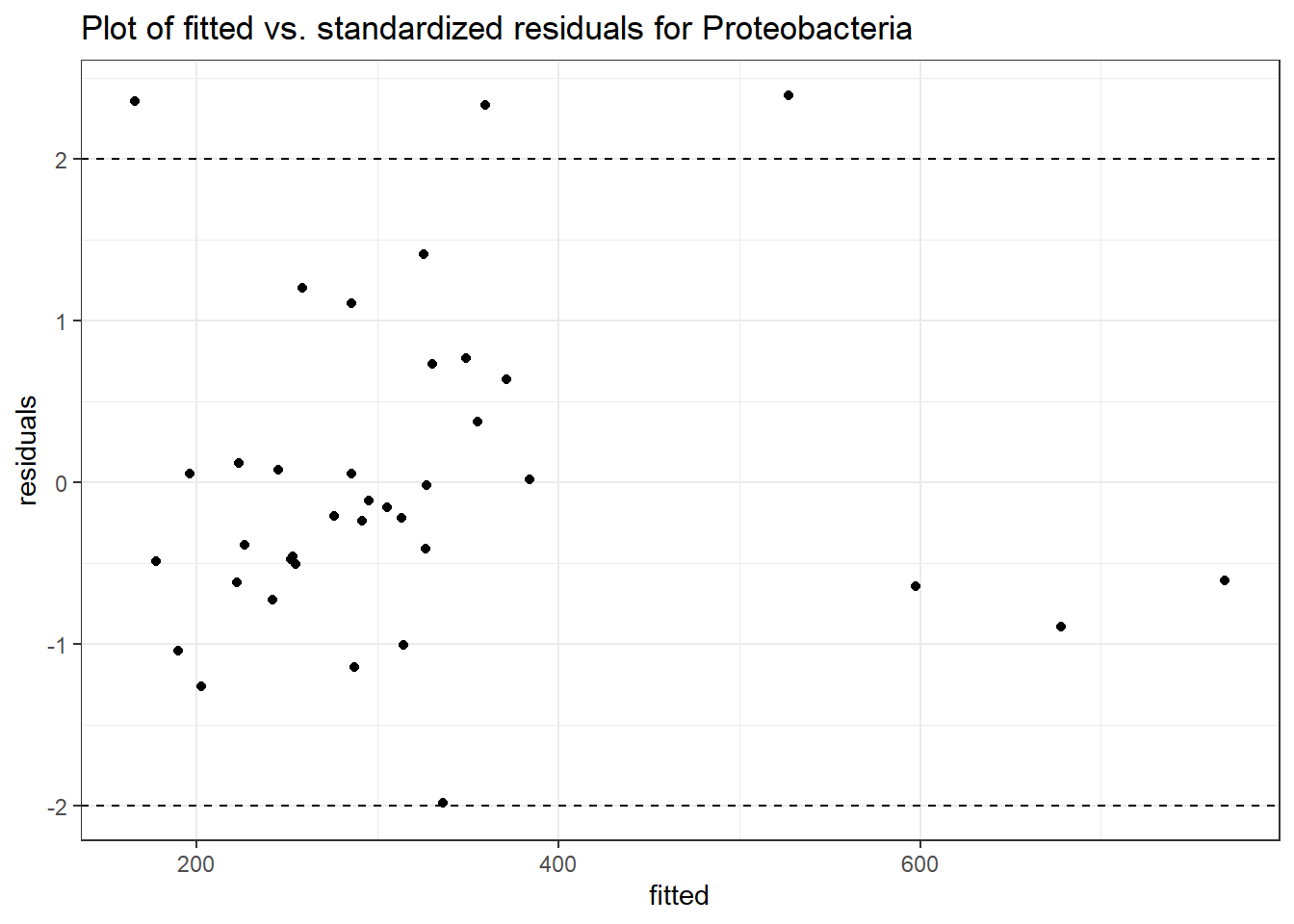

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.74272 0.10932 52.53091 0
time Proteobacteria -0.12581 0.01087 -11.56941 0
time:intB Proteobacteria 0.19257 0.01723 11.17407 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.35922
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.9209168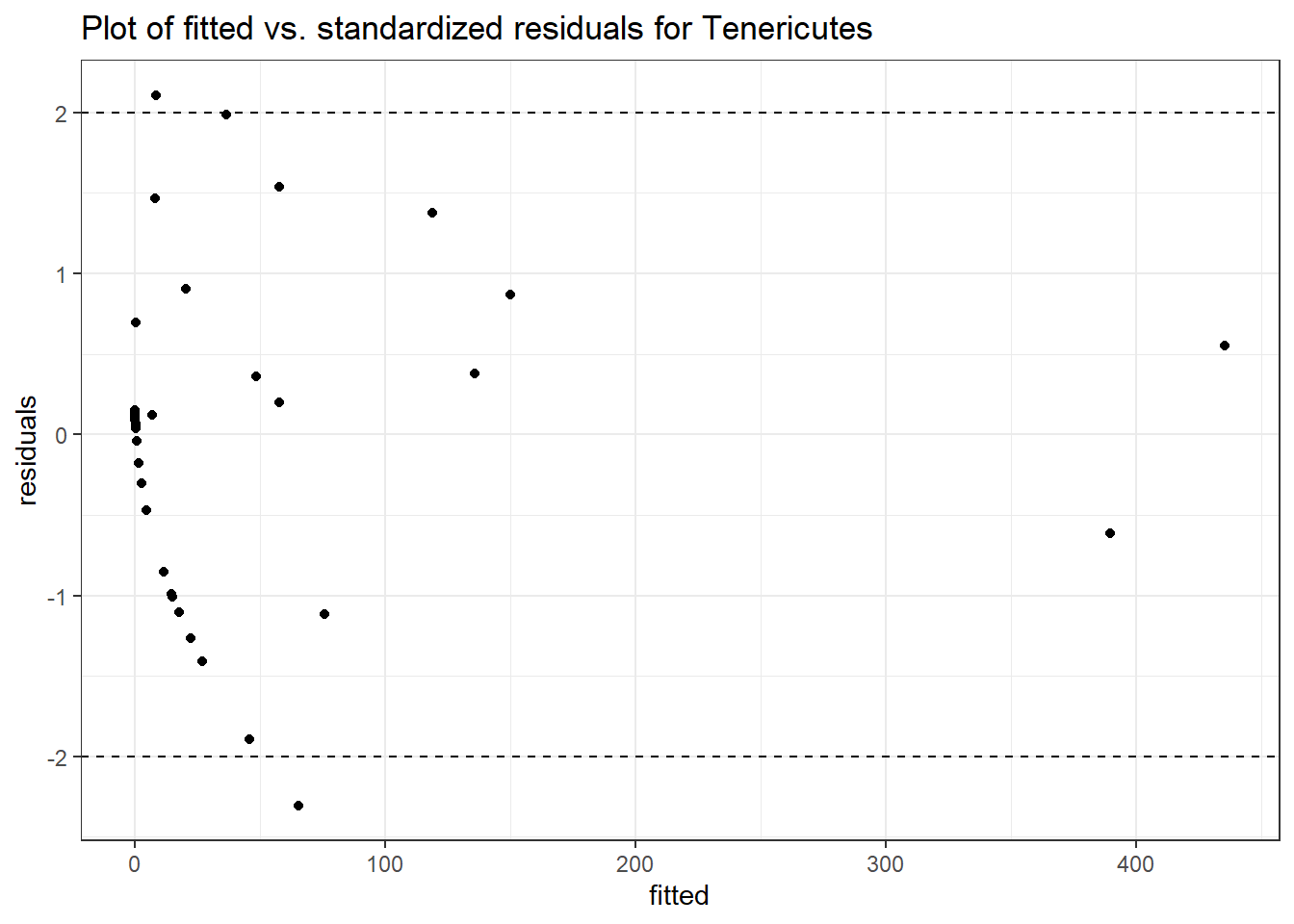
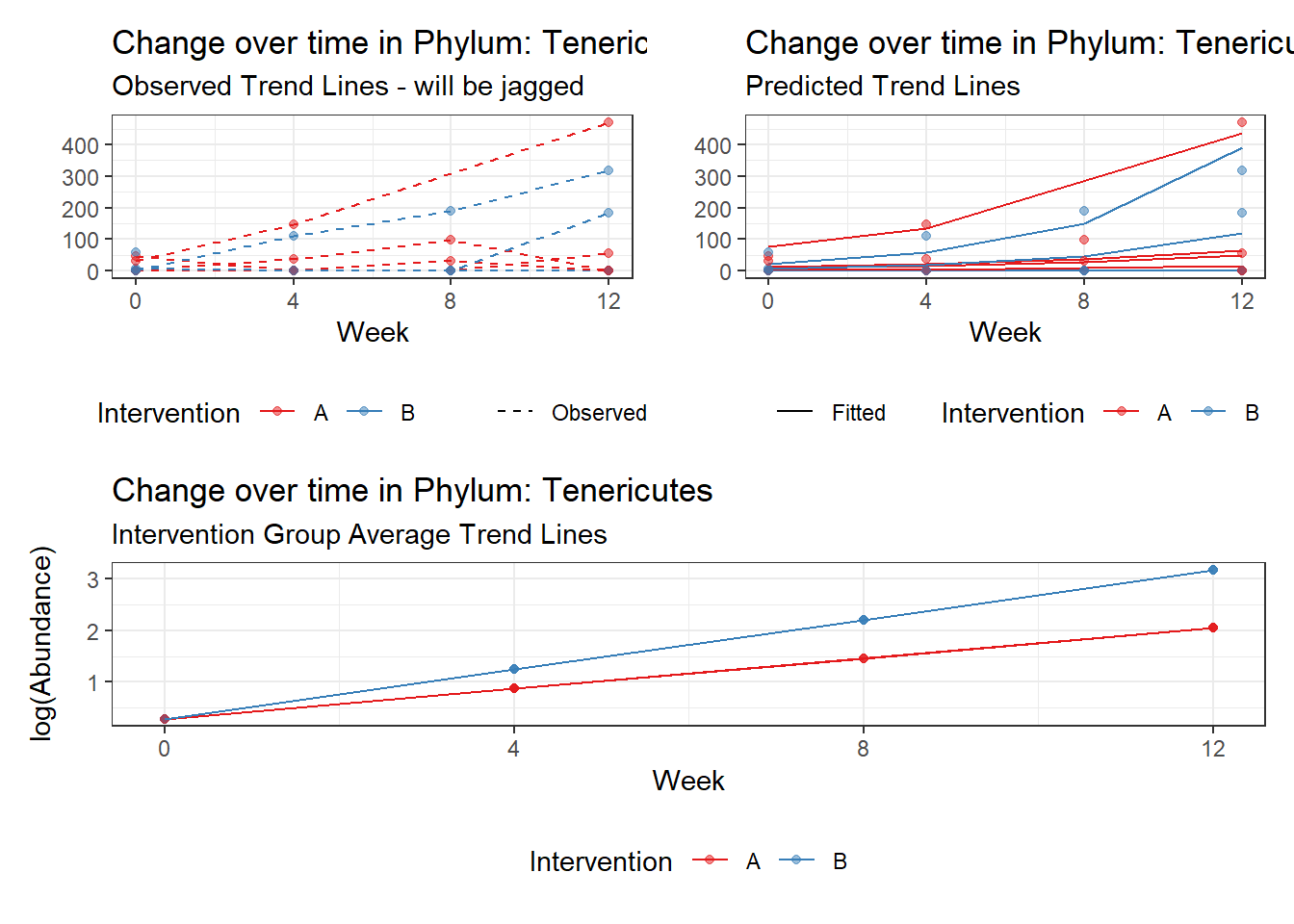
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 0.29442 1.06903 0.27541 0.783
time Tenericutes 0.58340 0.03023 19.29717 0.000
time:intB Tenericutes 0.37108 0.05290 7.01497 0.000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.4125
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000004cb03058>
Link: poisson
Intraclass Correlation (ICC): 0.8603956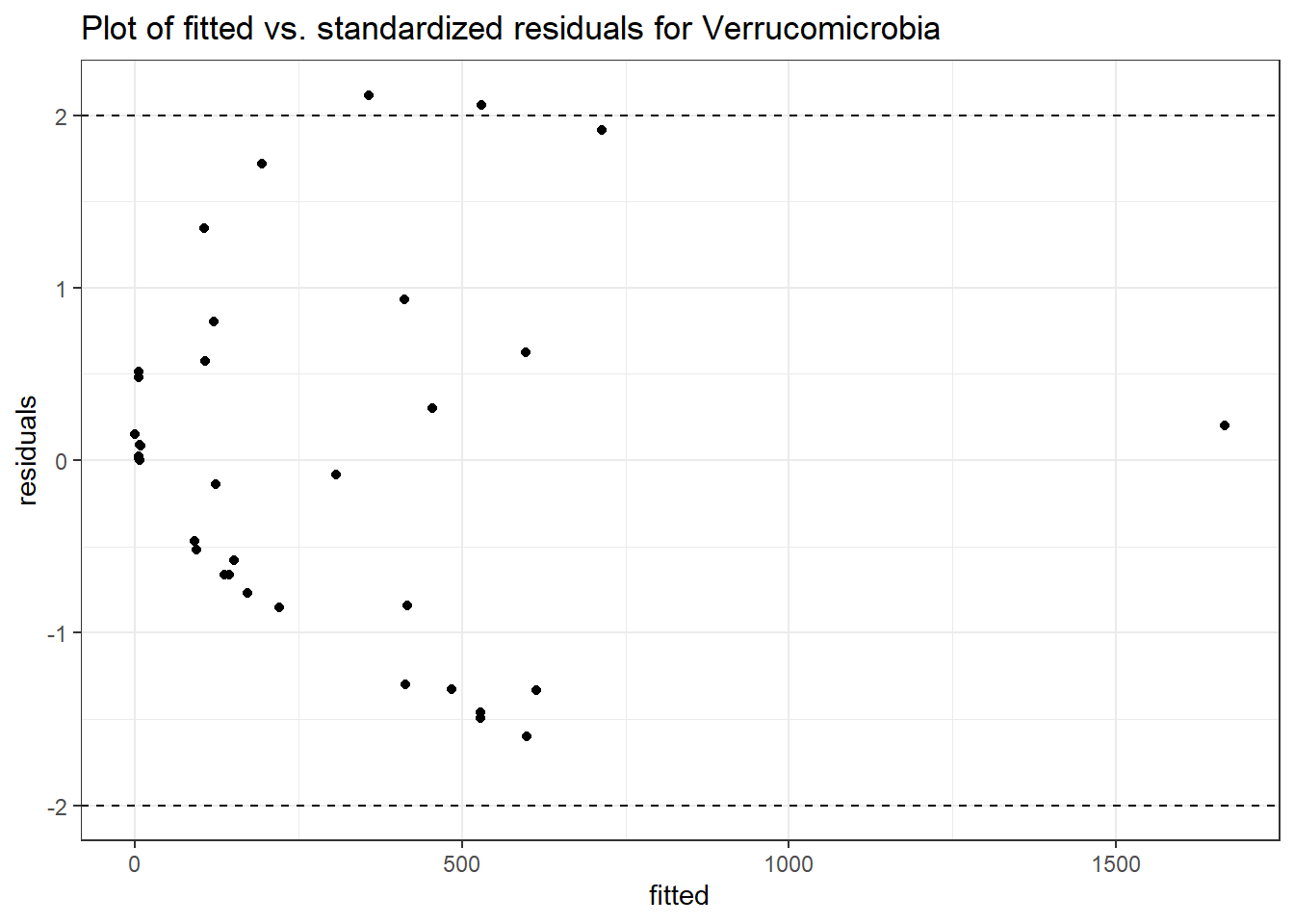
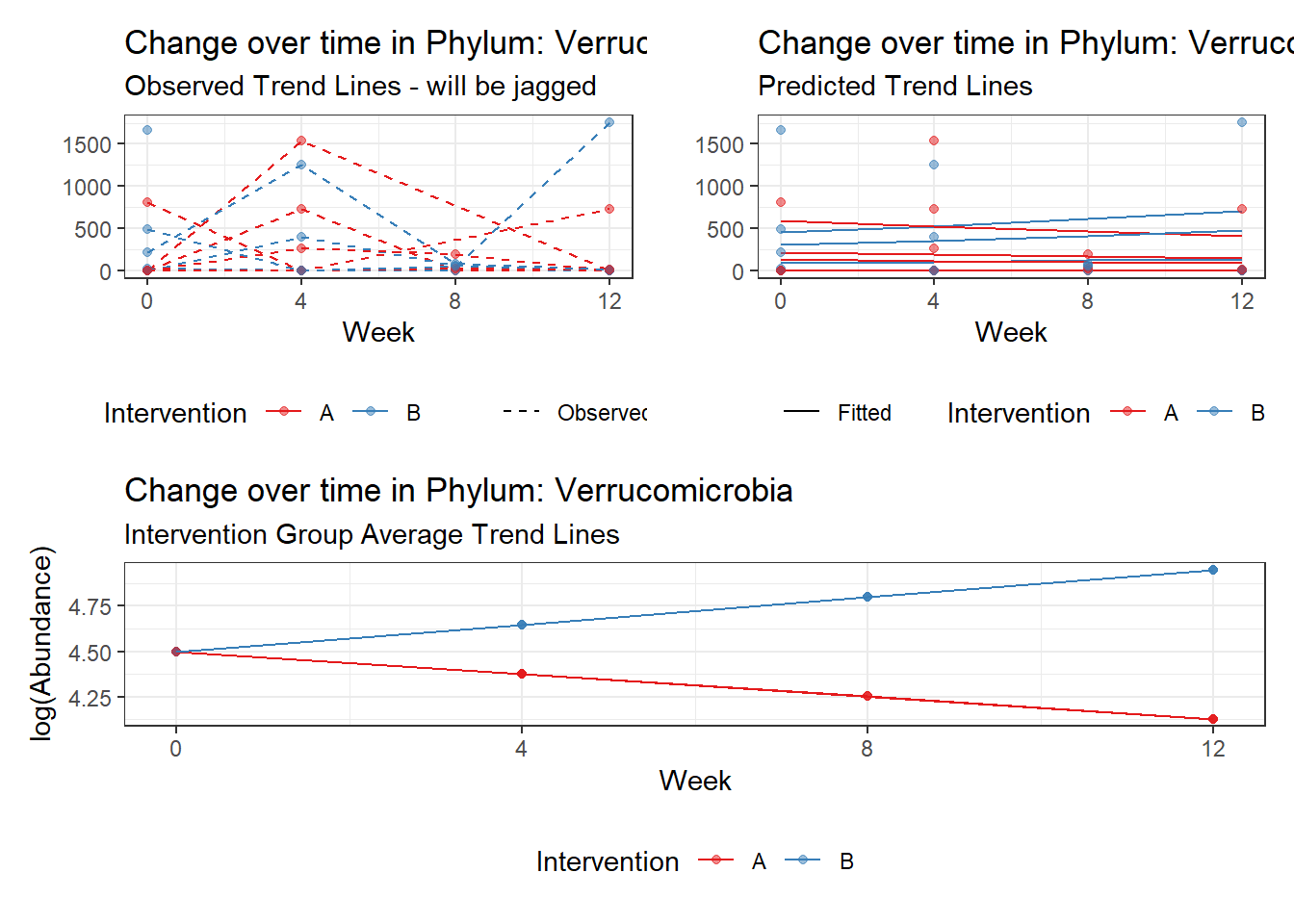
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.49776 0.75515 5.95610 0
time Verrucomicrobia -0.12318 0.01296 -9.50252 0
time:intB Verrucomicrobia 0.27351 0.01882 14.52988 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4826
Significance test not available for random effectsphylum.fit6b <- glmm_microbiome(mydata=microbiome_data,model.number=6,
taxa.level="Phylum", link="negbinom",
model="1 + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinom
Intraclass Correlation (ICC): 0.3183468
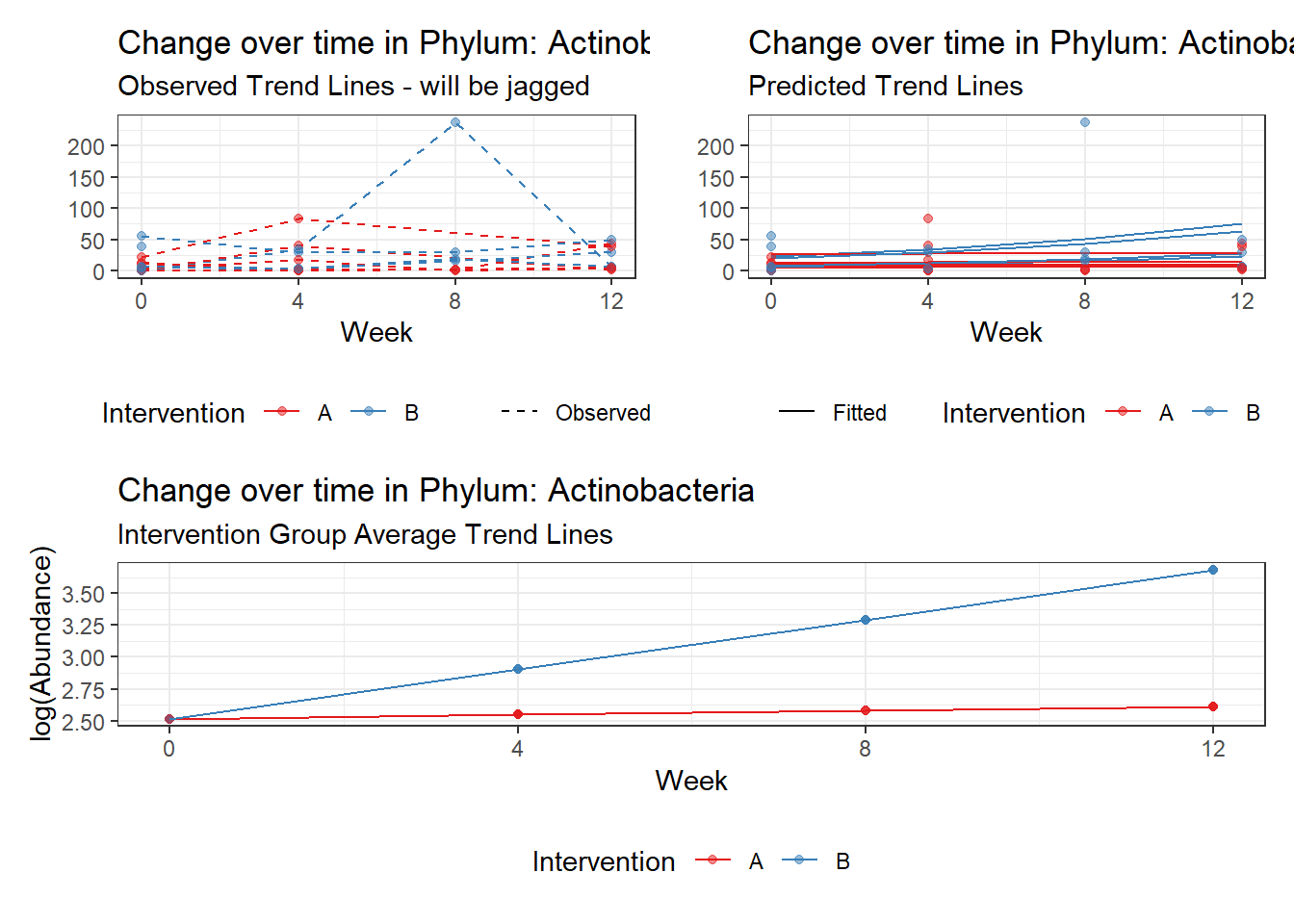
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.51648 0.39112 6.43396 0.00000
time Actinobacteria 0.03273 0.22700 0.14419 0.88535
time:intB Actinobacteria 0.35587 0.28469 1.25005 0.21128
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.68339
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinom
Intraclass Correlation (ICC): 0.1443309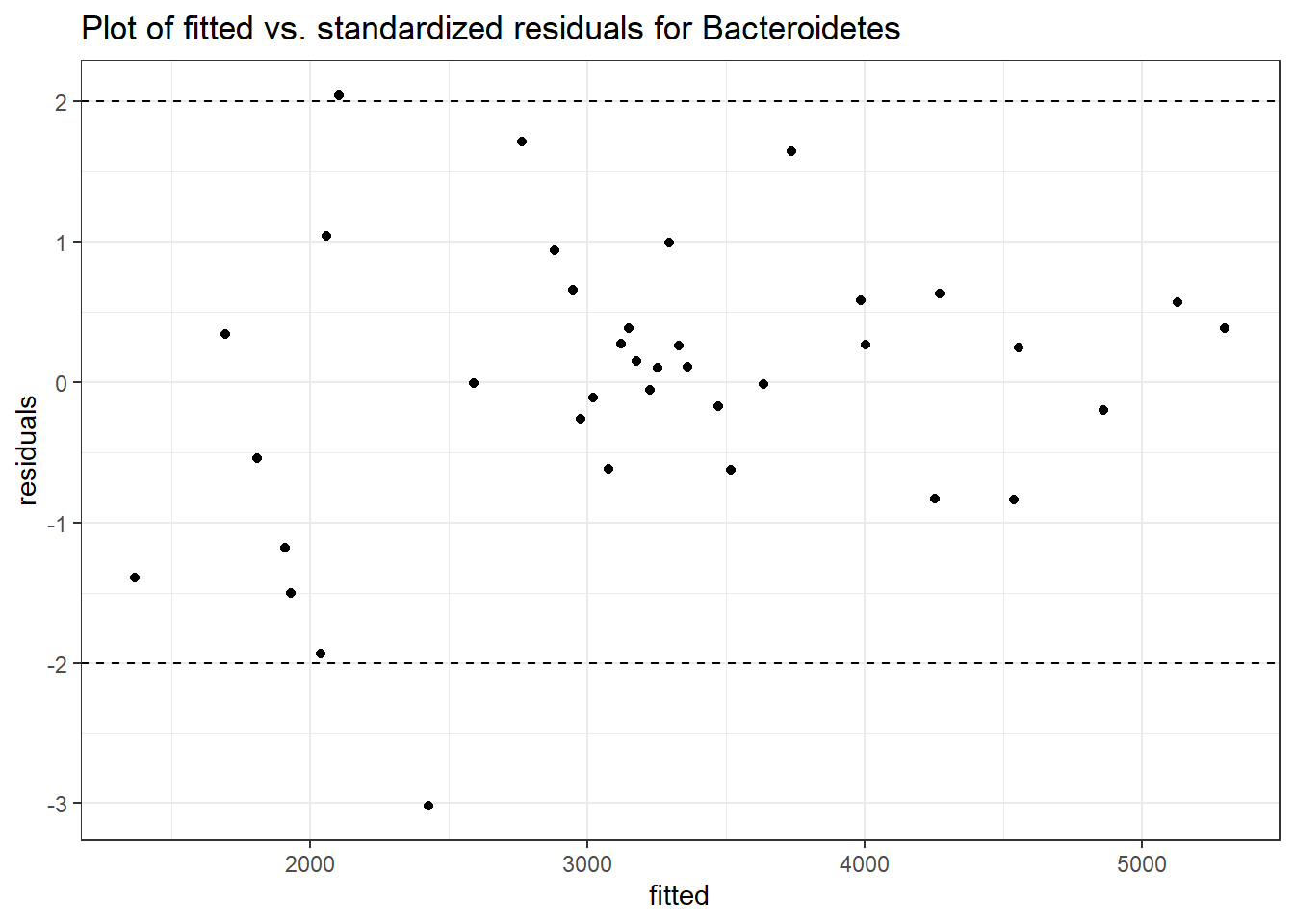
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.04560 0.13814 58.24076 0.00000
time Bacteroidetes -0.03257 0.04502 -0.72351 0.46937
time:intB Bacteroidetes -0.03216 0.06613 -0.48629 0.62676
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.4107
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced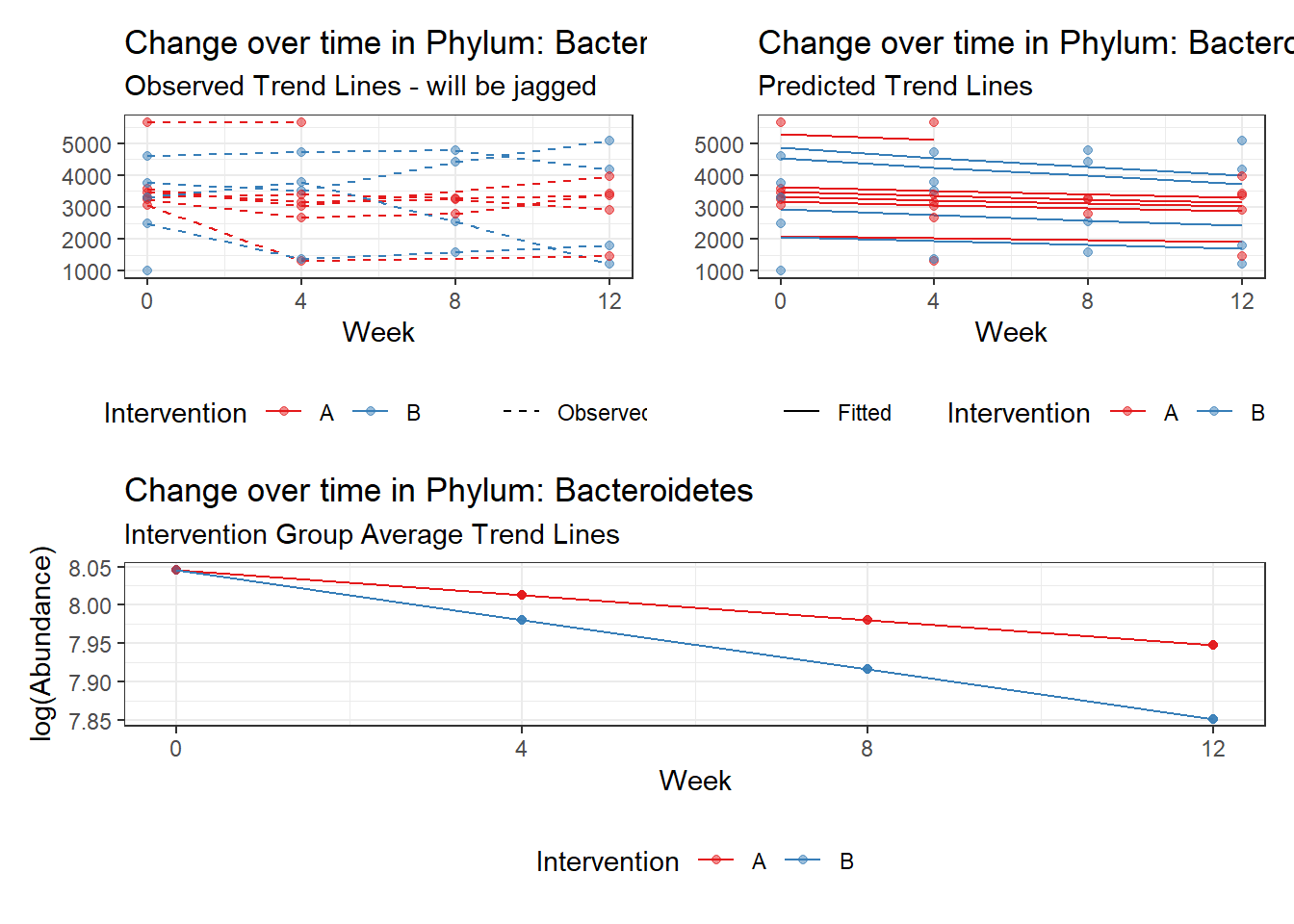
Intraclass Correlation (ICC): 0.9464282
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -0.86194 1.78254 -0.48355 0.62871
time Cyanobacteria -0.07268 0.04214 -1.72474 0.08457
time:intB Cyanobacteria -0.50410 0.06903 -7.30215 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.2032
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinomboundary (singular) fit: see ?isSingular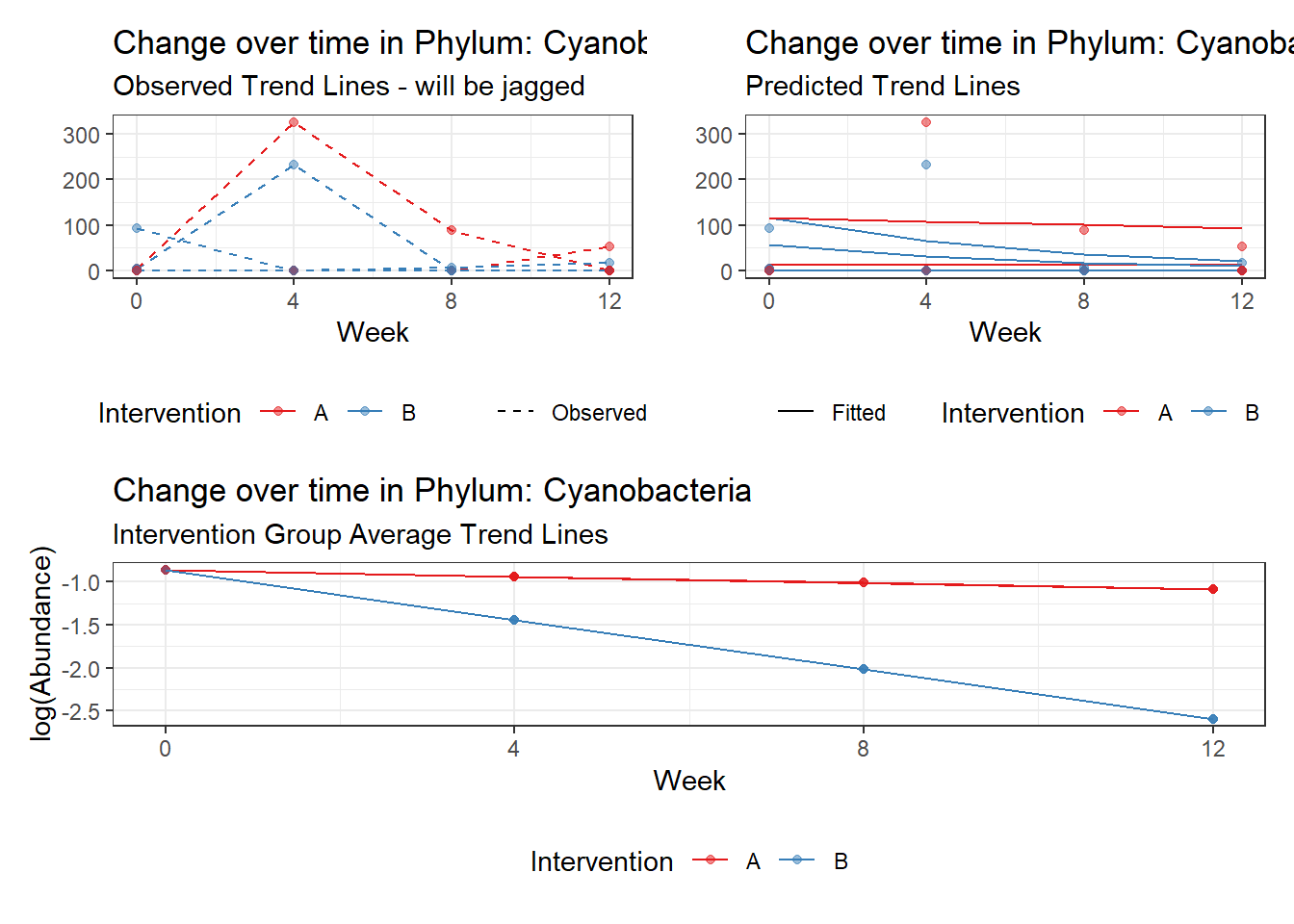
Intraclass Correlation (ICC): 8.624806e-12

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota 0.37213 0.74974 0.49634 0.61965
time Euryarchaeota -0.83611 0.57406 -1.45647 0.14526
time:intB Euryarchaeota 1.04268 0.61811 1.68688 0.09163
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.9368e-06
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinom
Intraclass Correlation (ICC): 0.1204993

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.92736 0.11904 66.59525 0.00000
time Firmicutes 0.03379 0.03205 1.05435 0.29172
time:intB Firmicutes -0.02624 0.04655 -0.56365 0.57300
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.37015
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinom
Intraclass Correlation (ICC): 0.8255539
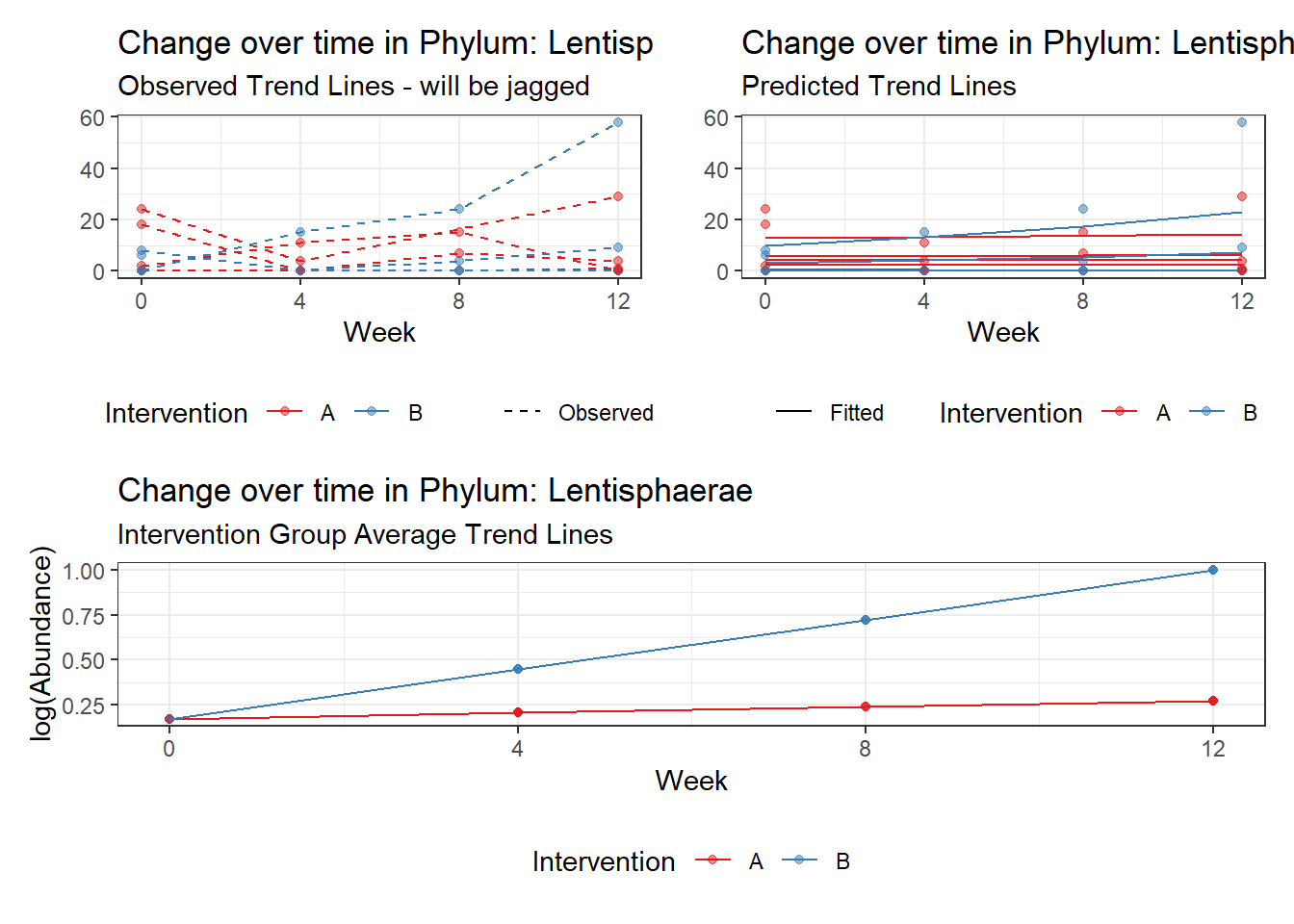
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.17260 0.93378 0.18484 0.85336
time Lentisphaerae 0.03298 0.36281 0.09089 0.92758
time:intB Lentisphaerae 0.24276 0.50880 0.47713 0.63327
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1754
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinom
Intraclass Correlation (ICC): 0.03896032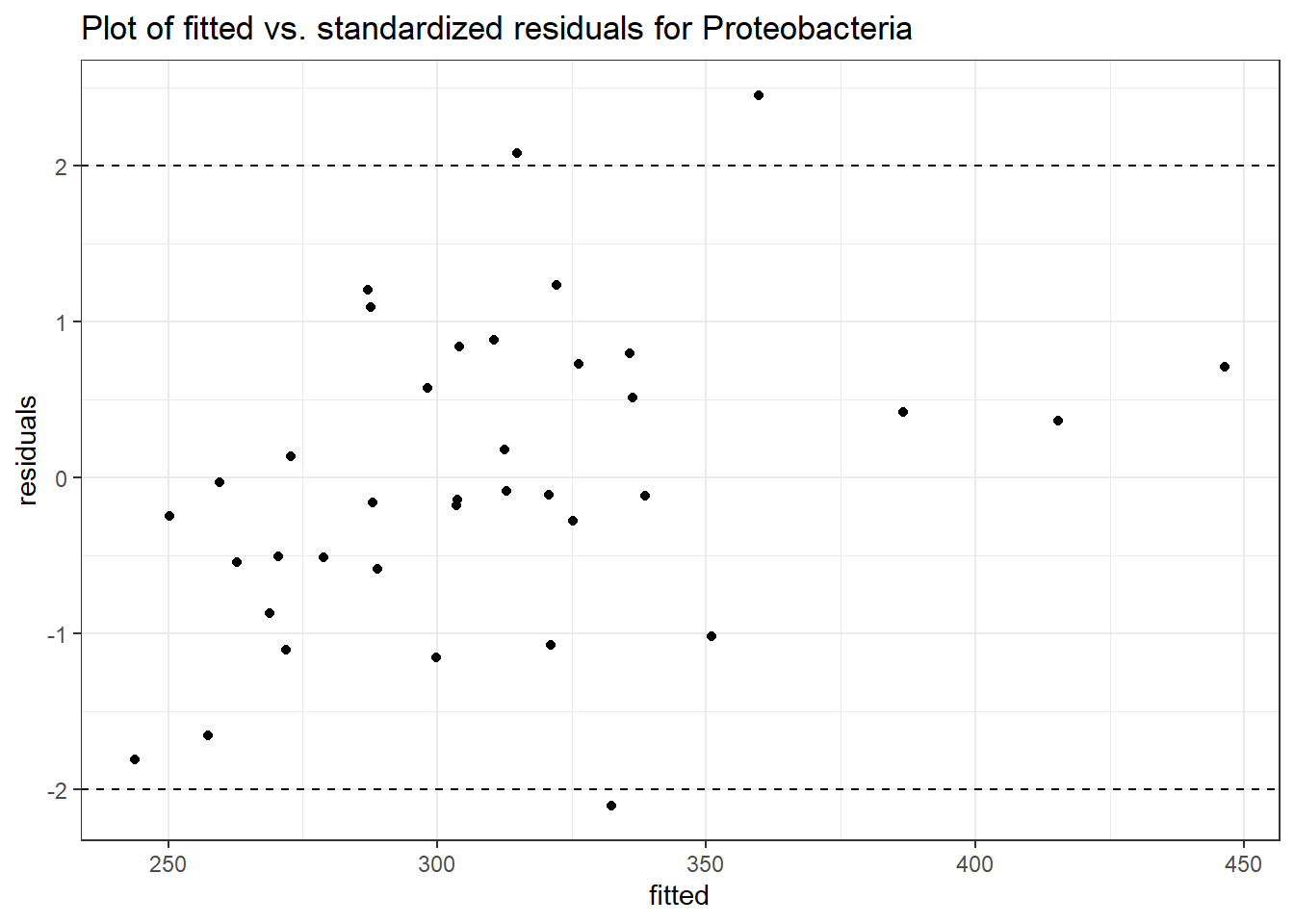
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.81254 0.16322 35.61208 0.00000
time Proteobacteria -0.07200 0.10958 -0.65702 0.51117
time:intB Proteobacteria 0.01761 0.13093 0.13446 0.89304
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.20134
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinomboundary (singular) fit: see ?isSingular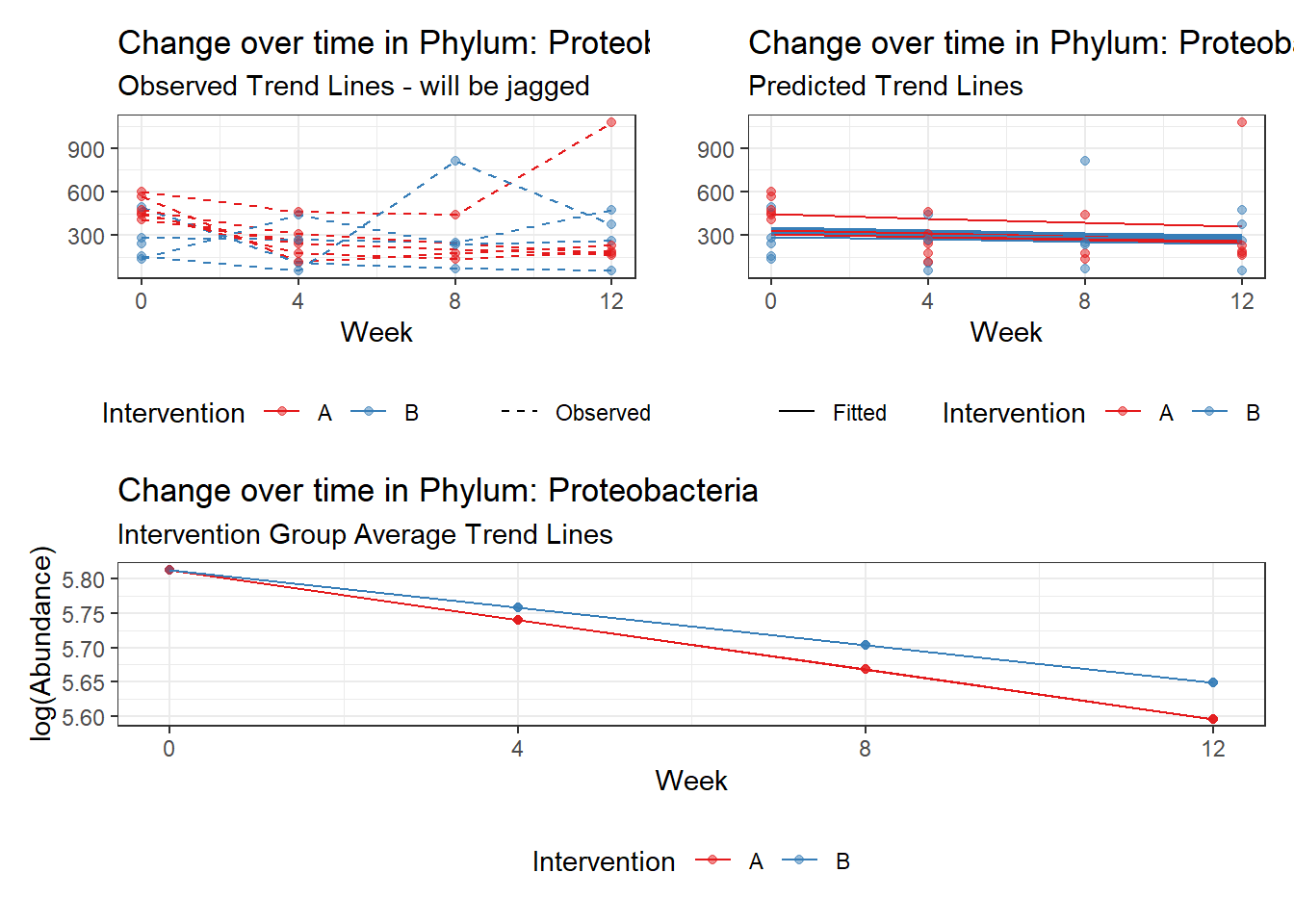
Intraclass Correlation (ICC): 6.796413e-11
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 2.57297 0.81255 3.16652 0.00154
time Tenericutes 0.69072 0.53347 1.29475 0.19541
time:intB Tenericutes 0.03490 0.57882 0.06029 0.95193
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 8.244e-06
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x000000000acec120>
Link: negbinomboundary (singular) fit: see ?isSingular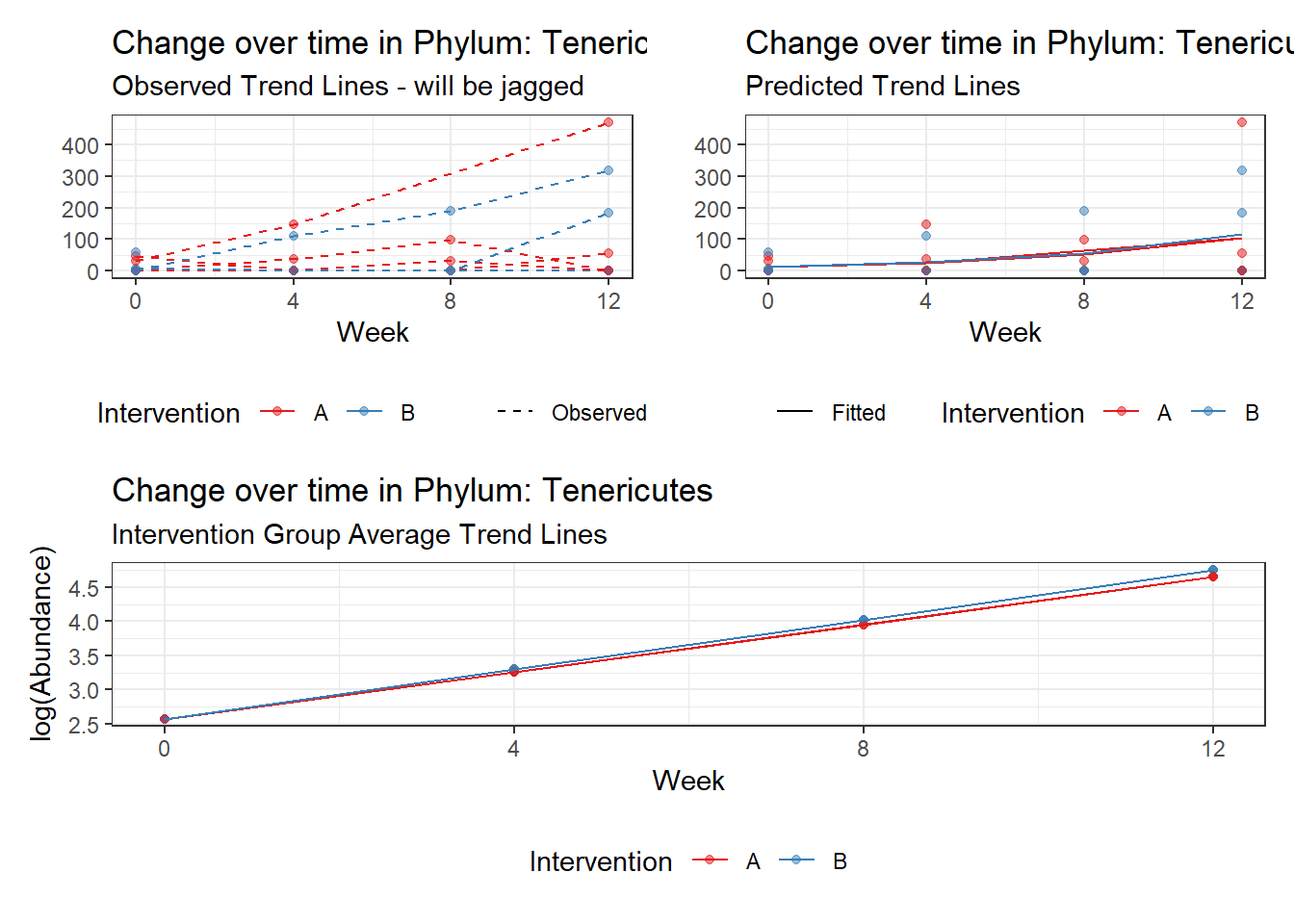
Intraclass Correlation (ICC): 8.057189e-12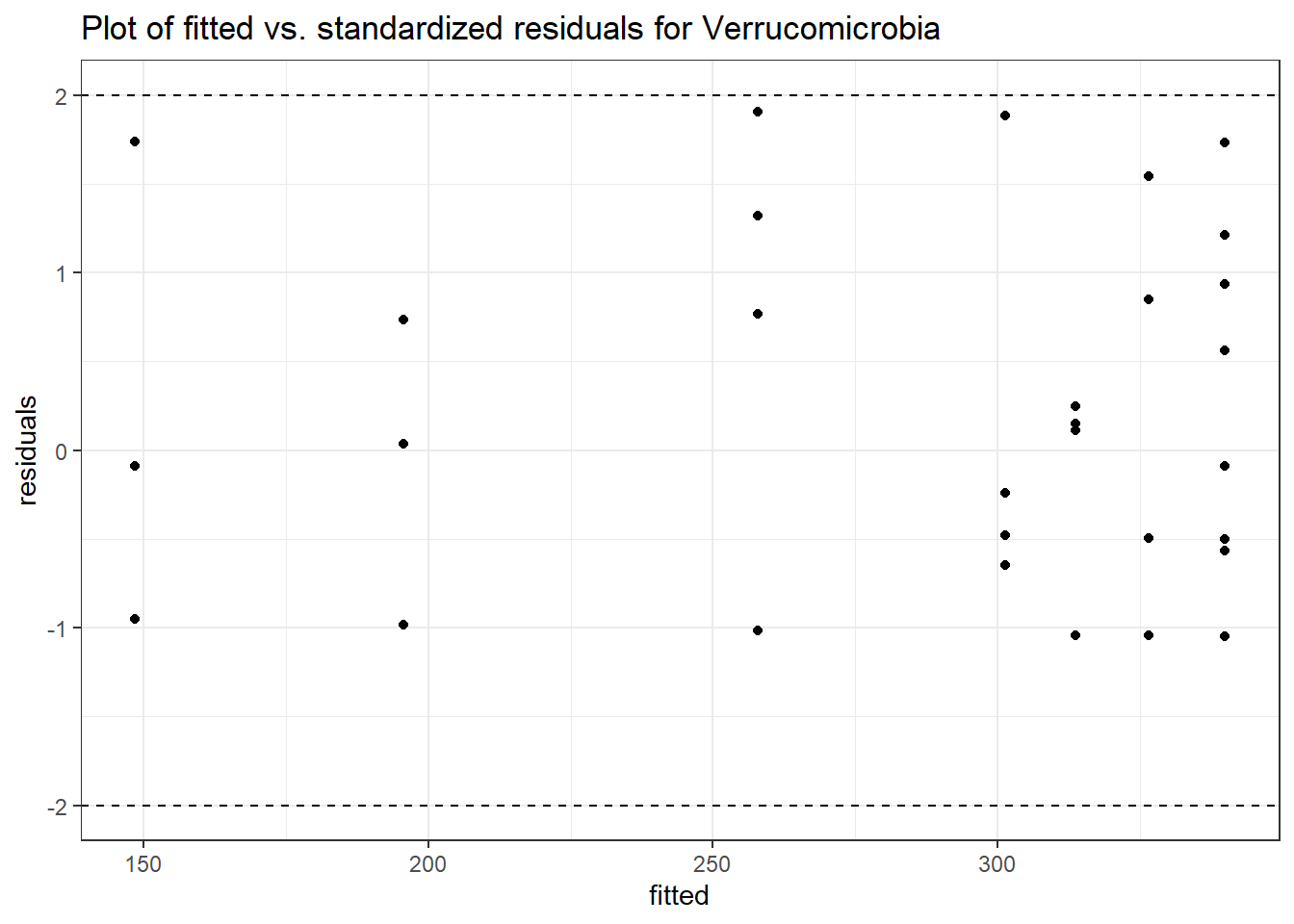
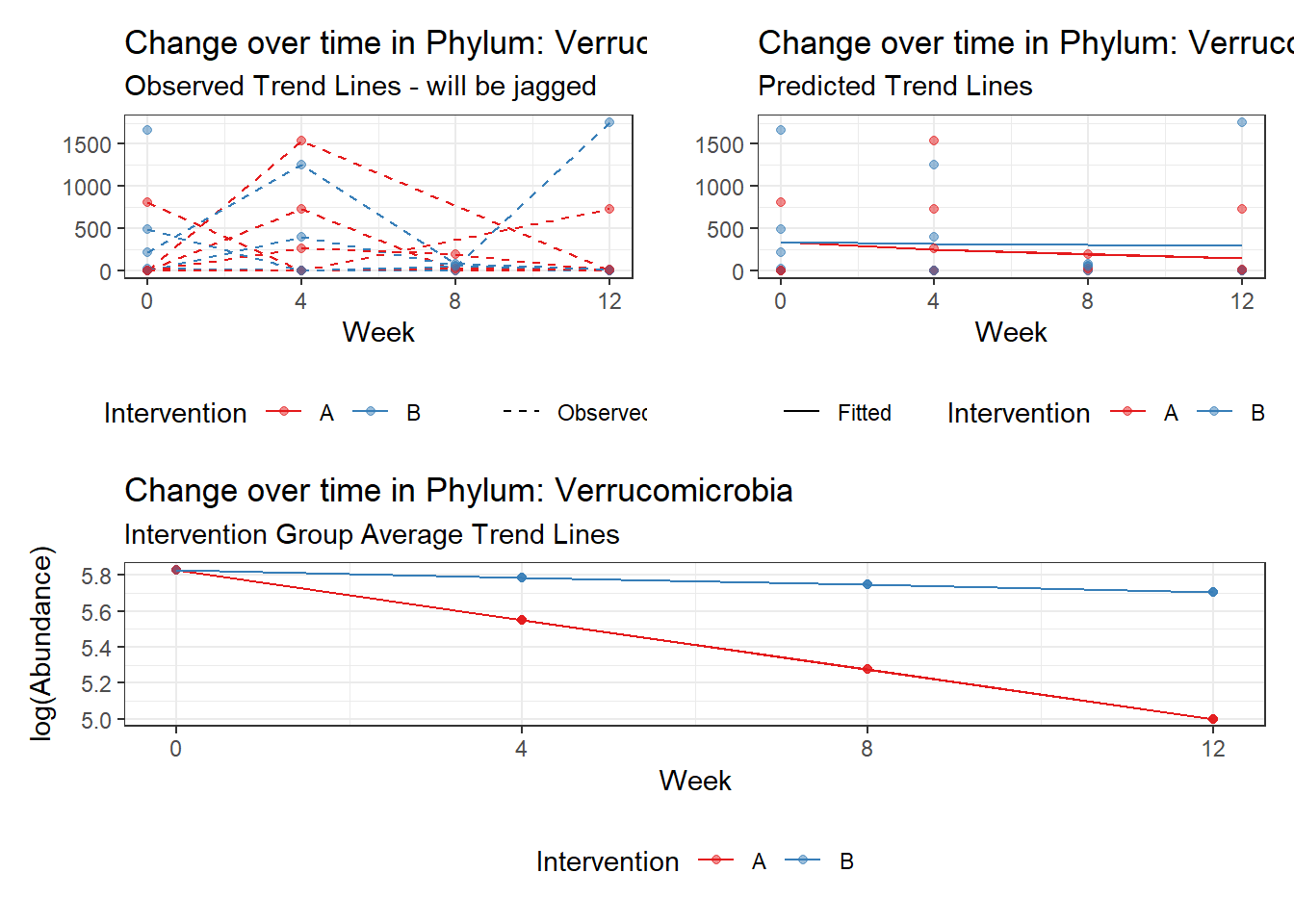
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 5.82888 0.65482 8.90148 0.00000
time Verrucomicrobia -0.27623 0.44654 -0.61860 0.53618
time:intB Verrucomicrobia 0.23601 0.46789 0.50442 0.61397
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.8385e-06
Significance test not available for random effectsCovariate(s): Time and Gender
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Gender)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
phylum.fit7 <- glmm_microbiome(mydata=microbiome_data, model.number=7,
taxa.level="Phylum", link="poisson",
model="1 + time + intB:time + female + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poisson
Intraclass Correlation (ICC): 0.4775383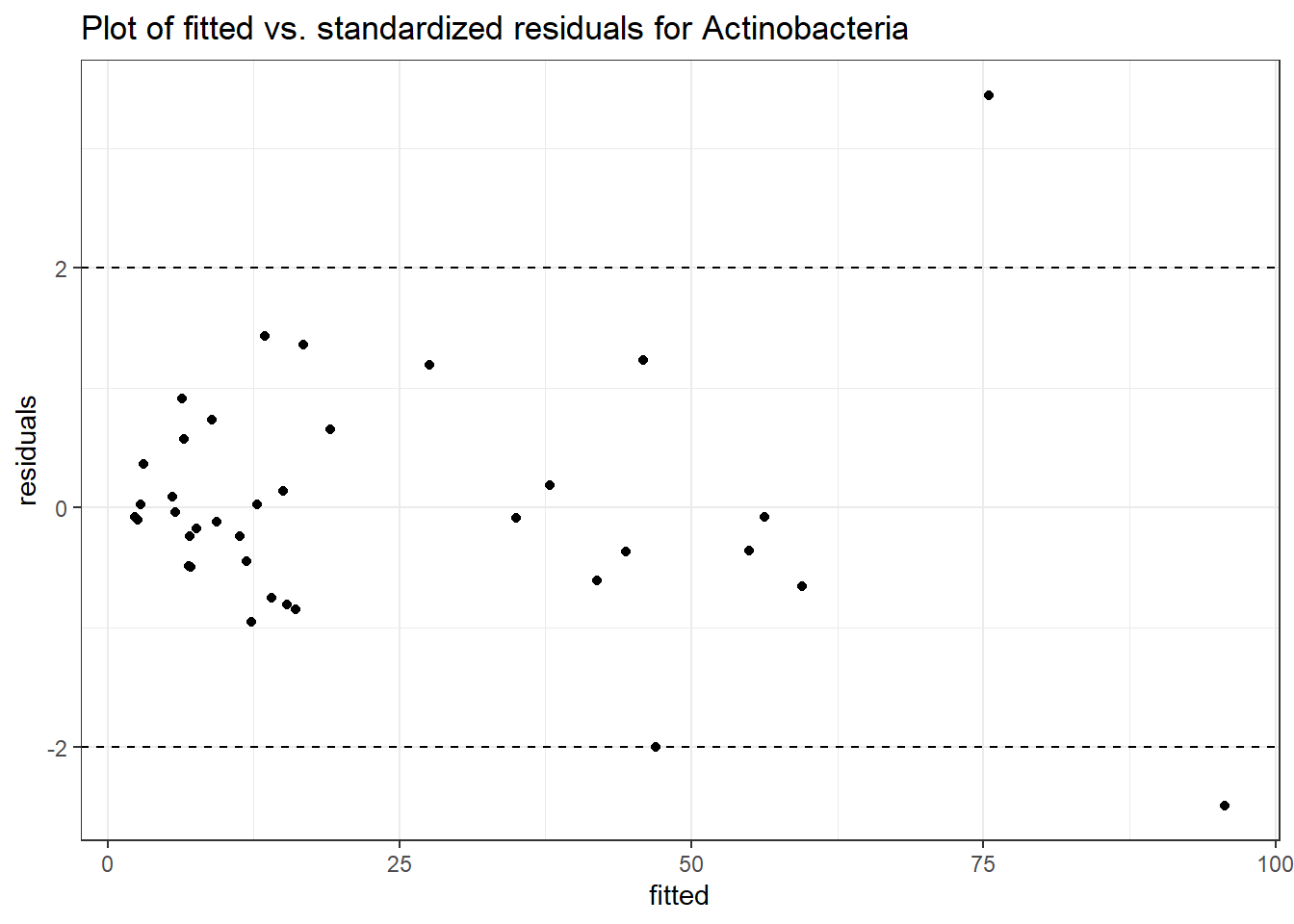
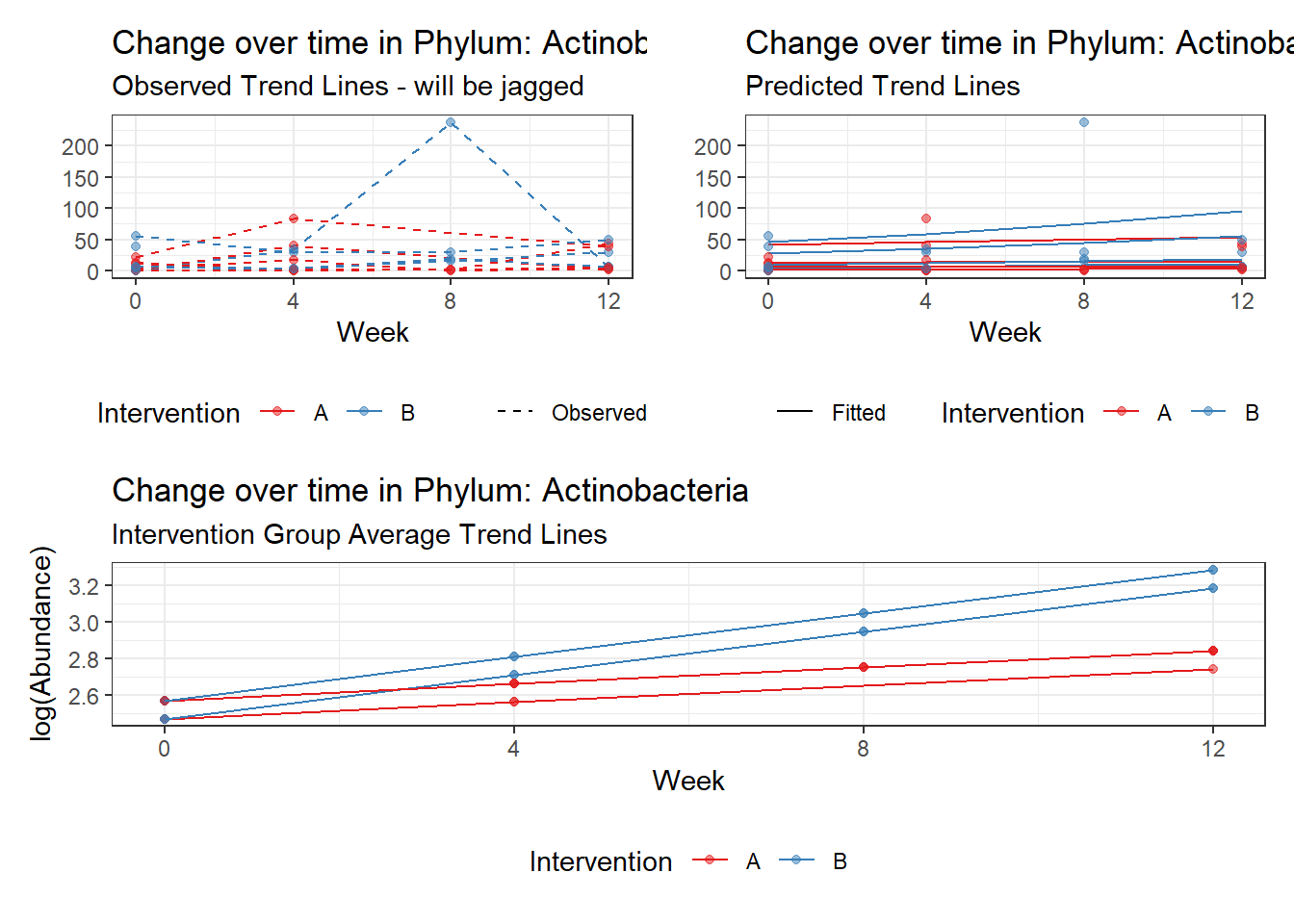
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.47224 0.48834 5.06255 0.00000
time Actinobacteria 0.09026 0.04833 1.86767 0.06181
female Actinobacteria 0.10018 0.60971 0.16431 0.86949
time:intB Actinobacteria 0.14736 0.06217 2.37020 0.01778
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.95604
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poisson
Intraclass Correlation (ICC): 0.1040945
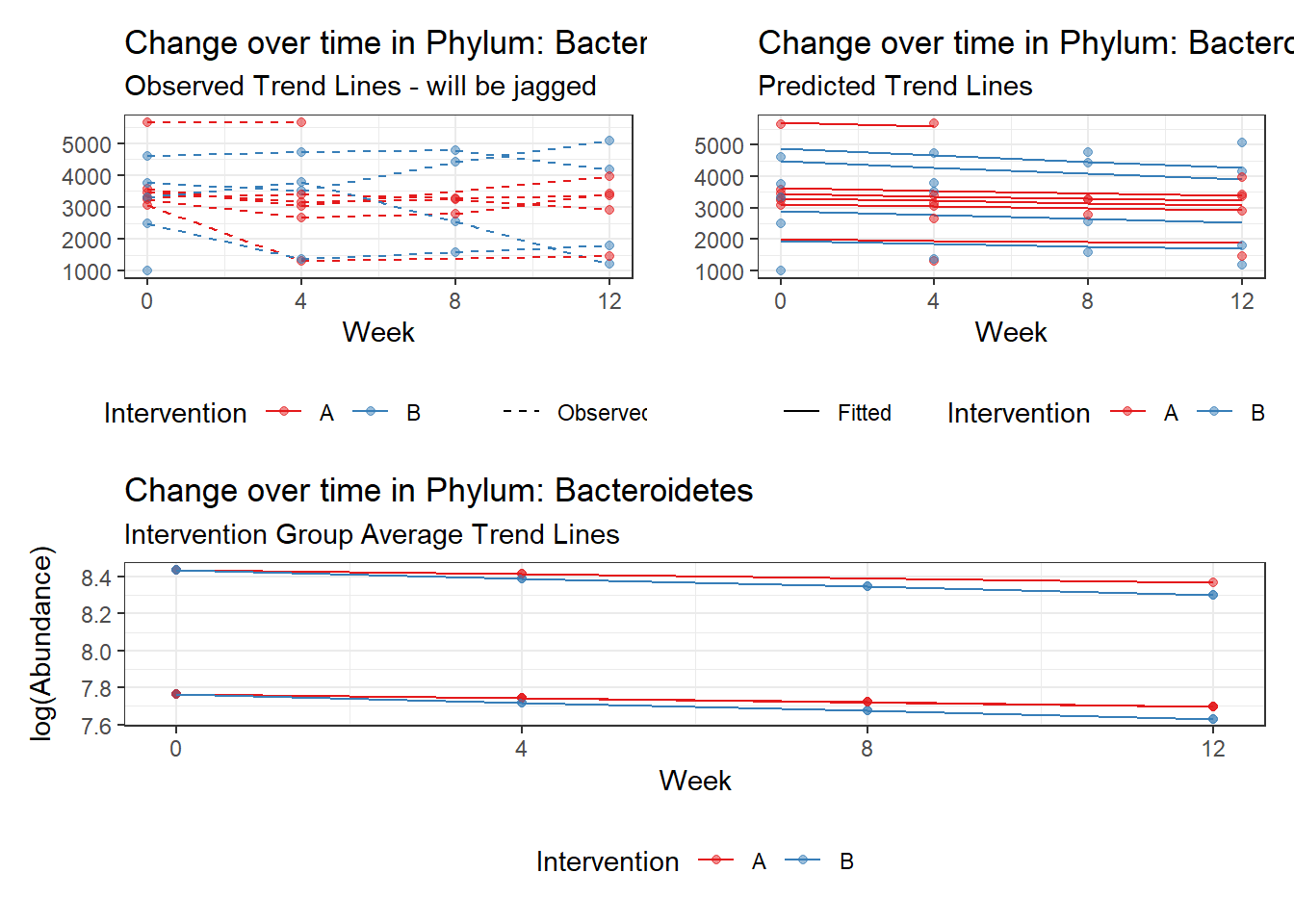
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.43685 0.17052 49.47786 0.00000
time Bacteroidetes -0.02172 0.00363 -5.98163 0.00000
female Bacteroidetes -0.67250 0.21378 -3.14578 0.00166
time:intB Bacteroidetes -0.02260 0.00531 -4.25457 0.00002
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.34087
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poisson
Intraclass Correlation (ICC): 0.9508193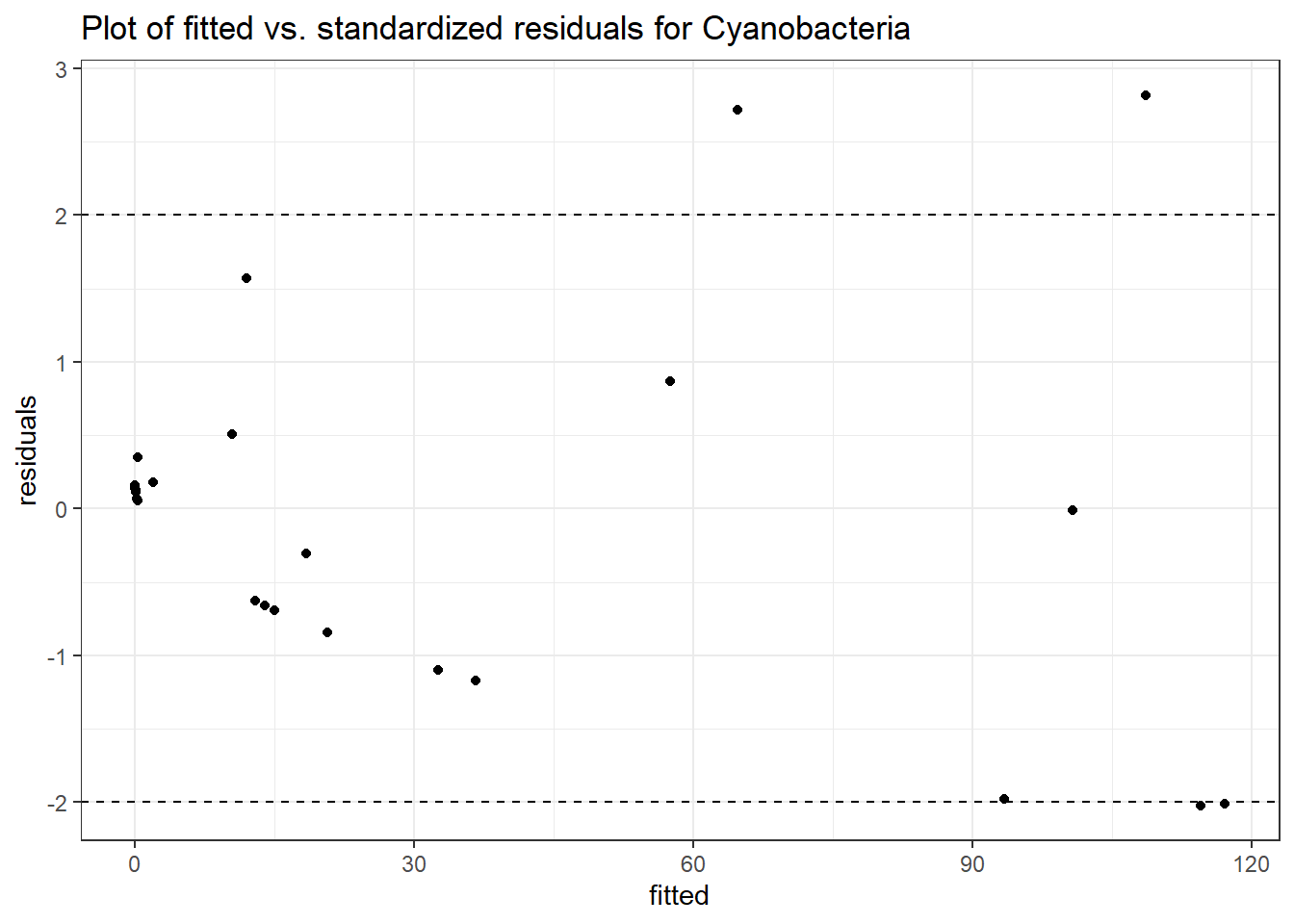
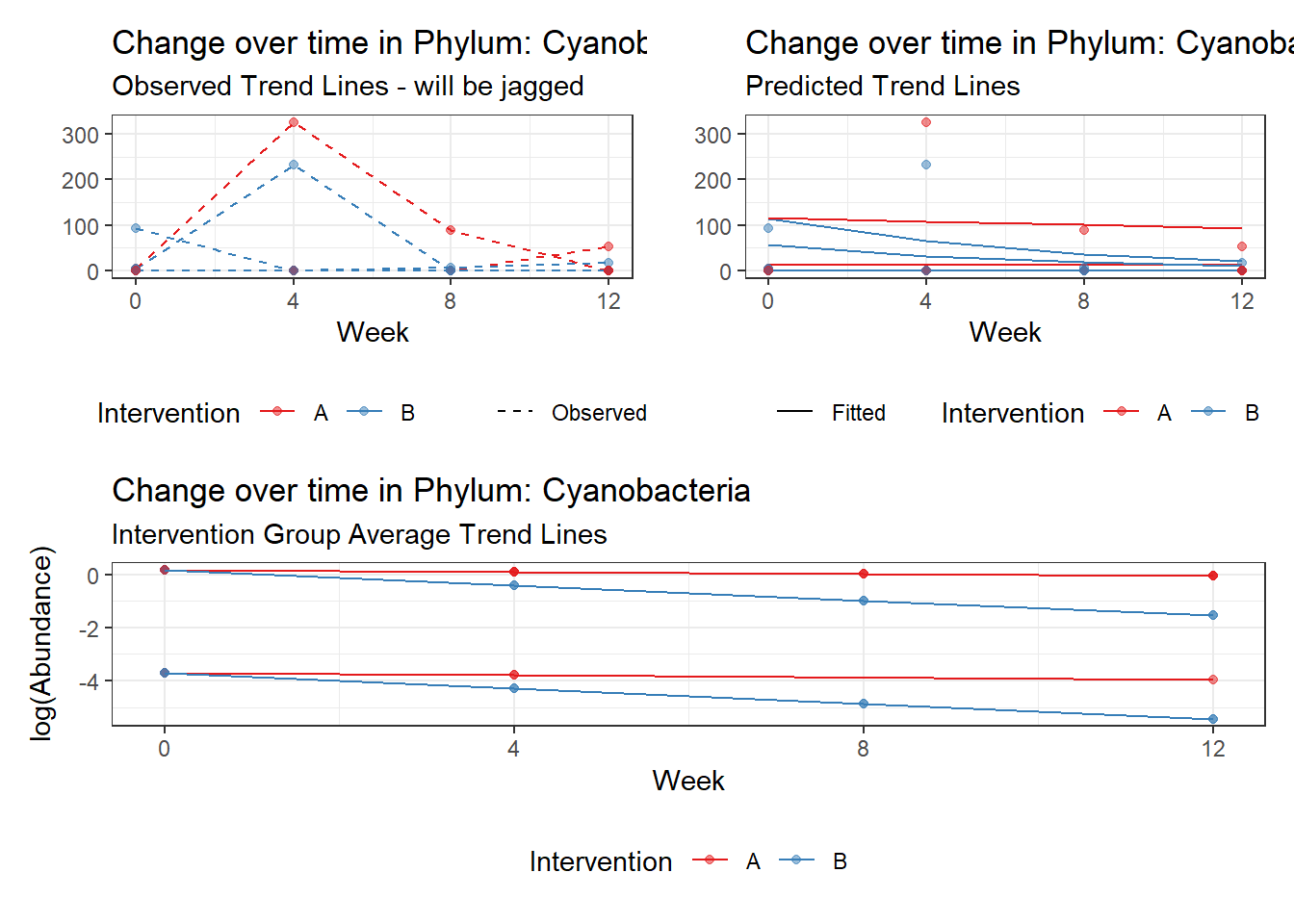
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -3.72197 3.18000 -1.17043 0.24183
time Cyanobacteria -0.07536 0.04112 -1.83249 0.06688
female Cyanobacteria 3.88290 3.35921 1.15590 0.24772
time:intB Cyanobacteria -0.49441 0.06784 -7.28822 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.3969
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poisson
Intraclass Correlation (ICC): 0.6687562

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -2.39191 1.28250 -1.86504 0.06218
time Euryarchaeota -0.35885 0.40875 -0.87793 0.37998
female Euryarchaeota 2.45898 1.27554 1.92779 0.05388
time:intB Euryarchaeota 0.39324 0.44285 0.88797 0.37455
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4209
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poisson
Intraclass Correlation (ICC): 0.05870396
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.52585 0.12513 60.14556 0e+00
time Firmicutes 0.03449 0.00352 9.79702 0e+00
female Firmicutes 0.62236 0.15675 3.97033 7e-05
time:intB Firmicutes -0.01847 0.00538 -3.43189 6e-04
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.24973
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poissonWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
unable to evaluate scaled gradientWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge: degenerate Hessian with 1 negative eigenvalues
Intraclass Correlation (ICC): 0.3584695
Warning in vcov.merMod(object, use.hessian = use.hessian): variance-covariance matrix computed from finite-difference Hessian is
not positive definite or contains NA values: falling back to var-cov estimated from RXWarning in vcov.merMod(object, correlation = correlation, sigm = sig): variance-covariance matrix computed from finite-difference Hessian is
not positive definite or contains NA values: falling back to var-cov estimated from RX
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae -19.14196 2412.87711 -0.00793 0.99367
time Lentisphaerae 0.00049 0.07813 0.00633 0.99495
female Lentisphaerae 20.76479 2412.87712 0.00861 0.99313
time:intB Lentisphaerae 0.69748 0.11882 5.87025 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.74751
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poisson
Intraclass Correlation (ICC): 0.1059356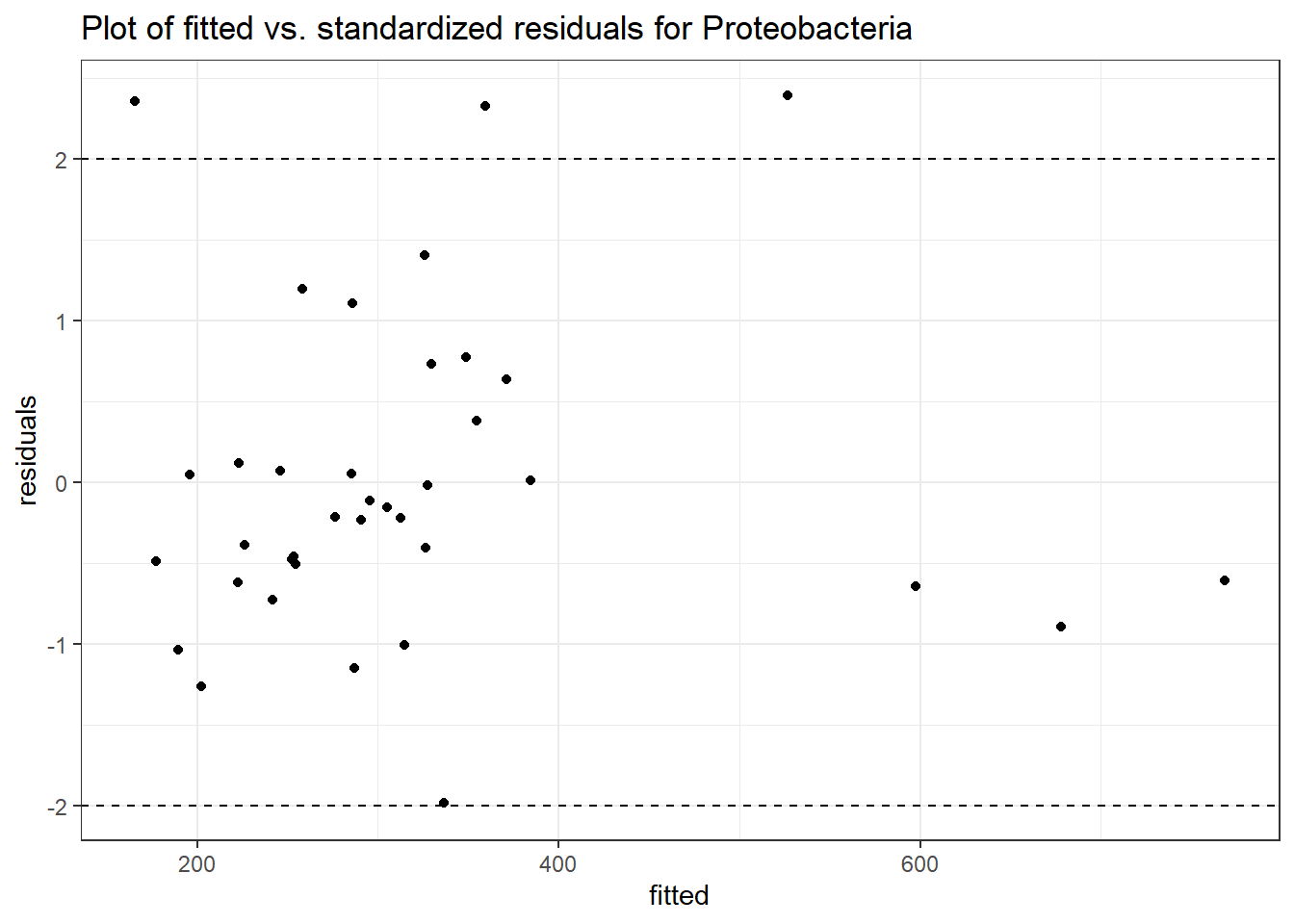

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.60737 0.17337 32.34425 0.00000
time Proteobacteria -0.12587 0.01087 -11.57691 0.00000
female Proteobacteria 0.21288 0.21695 0.98122 0.32648
time:intB Proteobacteria 0.19271 0.01723 11.18713 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.34422
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poisson
Intraclass Correlation (ICC): 0.5427045
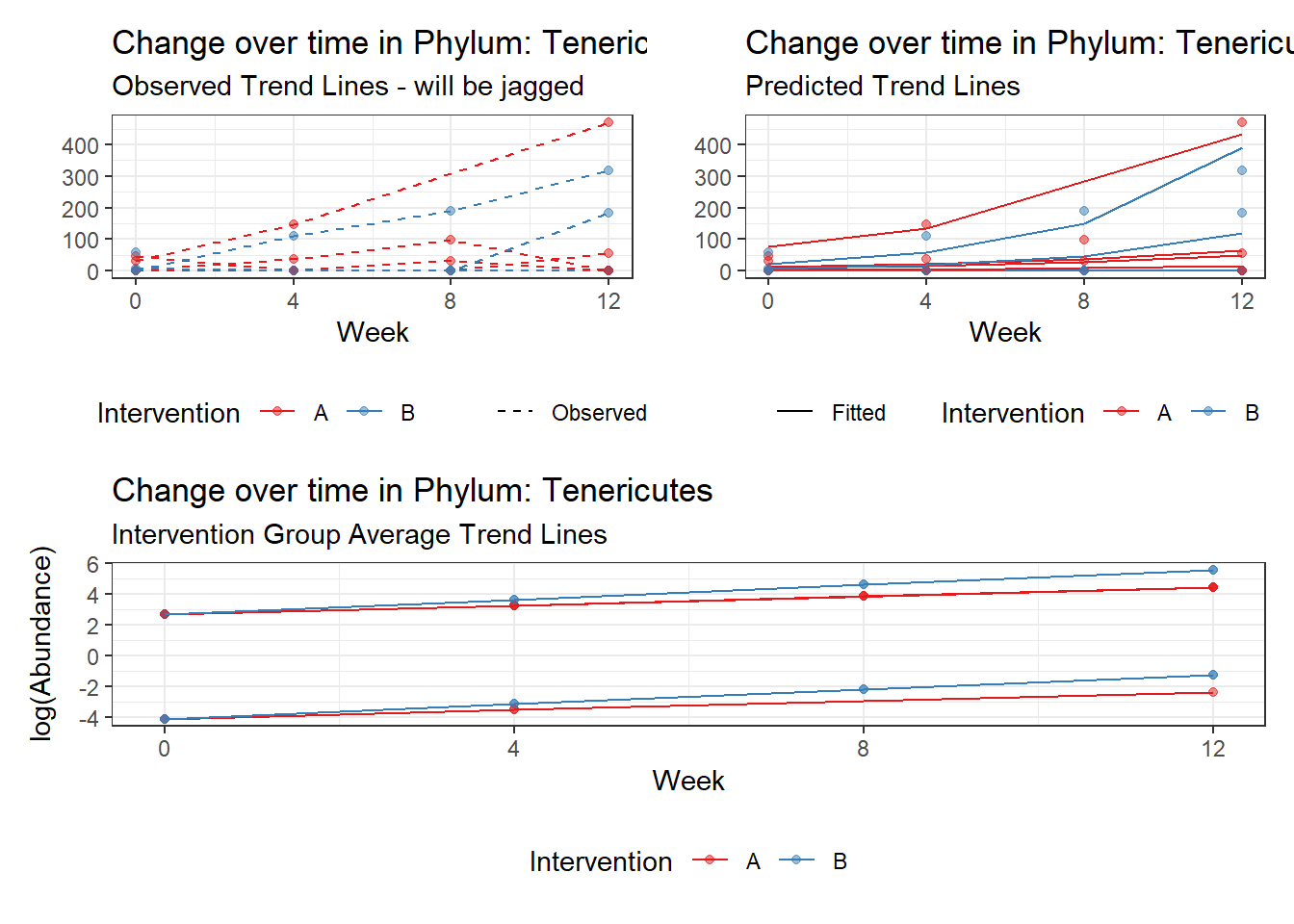
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes -4.08068 1.04917 -3.88942 1e-04
time Tenericutes 0.58101 0.03014 19.27542 0e+00
female Tenericutes 6.78313 1.12532 6.02774 0e+00
time:intB Tenericutes 0.37468 0.05251 7.13563 0e+00
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0894
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004aa9b788>
Link: poisson
Intraclass Correlation (ICC): 0.8231144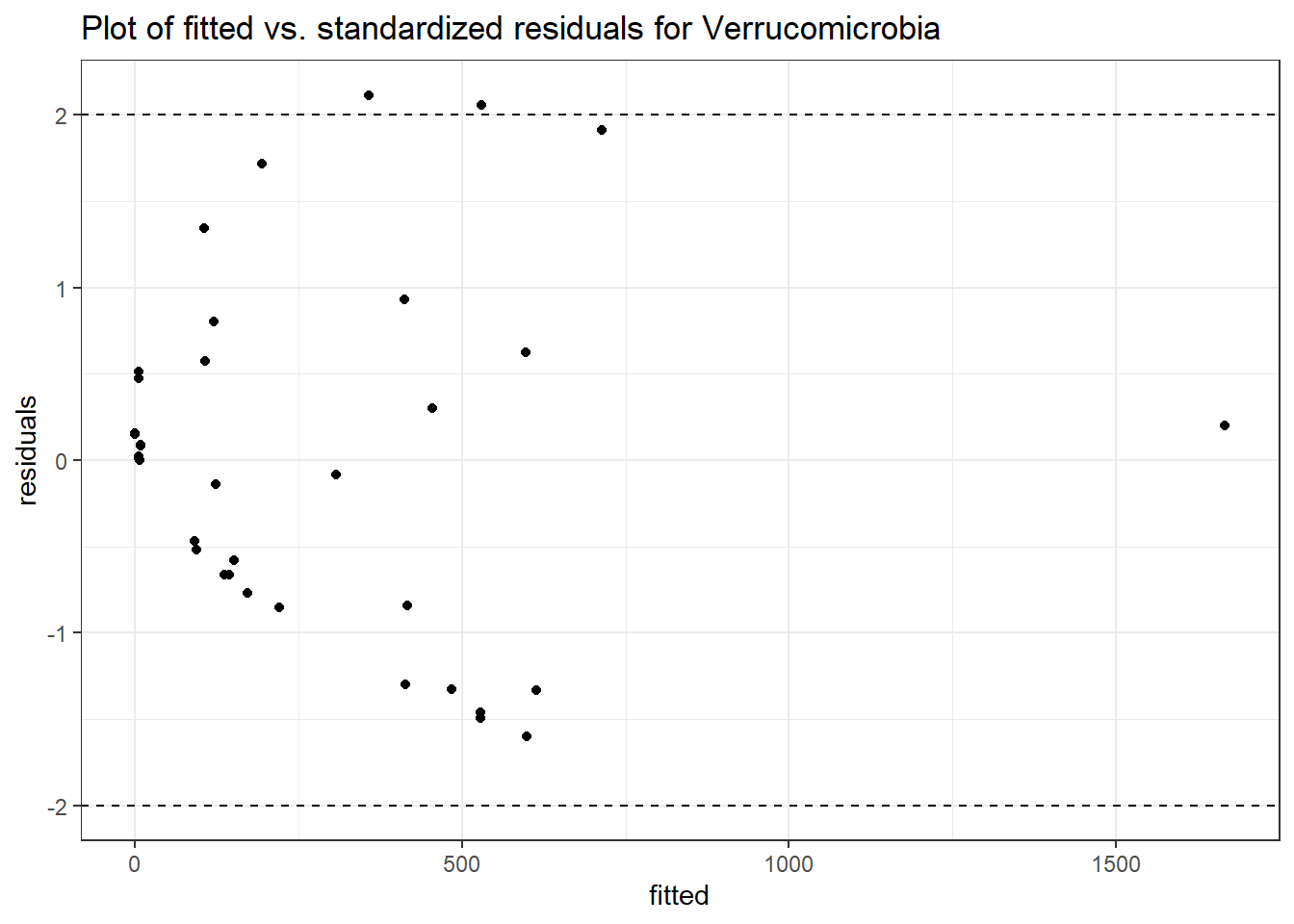

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 2.84073 1.11205 2.55450 0.01063
time Verrucomicrobia -0.12326 0.01296 -9.50863 0.00000
female Verrucomicrobia 2.59757 1.37874 1.88402 0.05956
time:intB Verrucomicrobia 0.27369 0.01882 14.53922 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1572
Significance test not available for random effectsphylum.fit7b <- glmm_microbiome(mydata=microbiome_data, model.number=7,
taxa.level="Phylum", link="negbinom",
model="1 + time + intB:time + female + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004d965328>
Link: negbinom
Intraclass Correlation (ICC): 0.317027
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.45484 0.55600 4.41515 0.00001
time Actinobacteria 0.02574 0.23155 0.11117 0.91149
female Actinobacteria 0.09776 0.62458 0.15653 0.87562
time:intB Actinobacteria 0.36913 0.29757 1.24048 0.21480
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.68131
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + intB:time + female + (1 | SubjectID)
<environment: 0x000000004d965328>
Link: negbinomError in factory(refitNB, types = c("message", "warning"))(lastfit, theta = exp(t), : Downdated VtV is not positive definite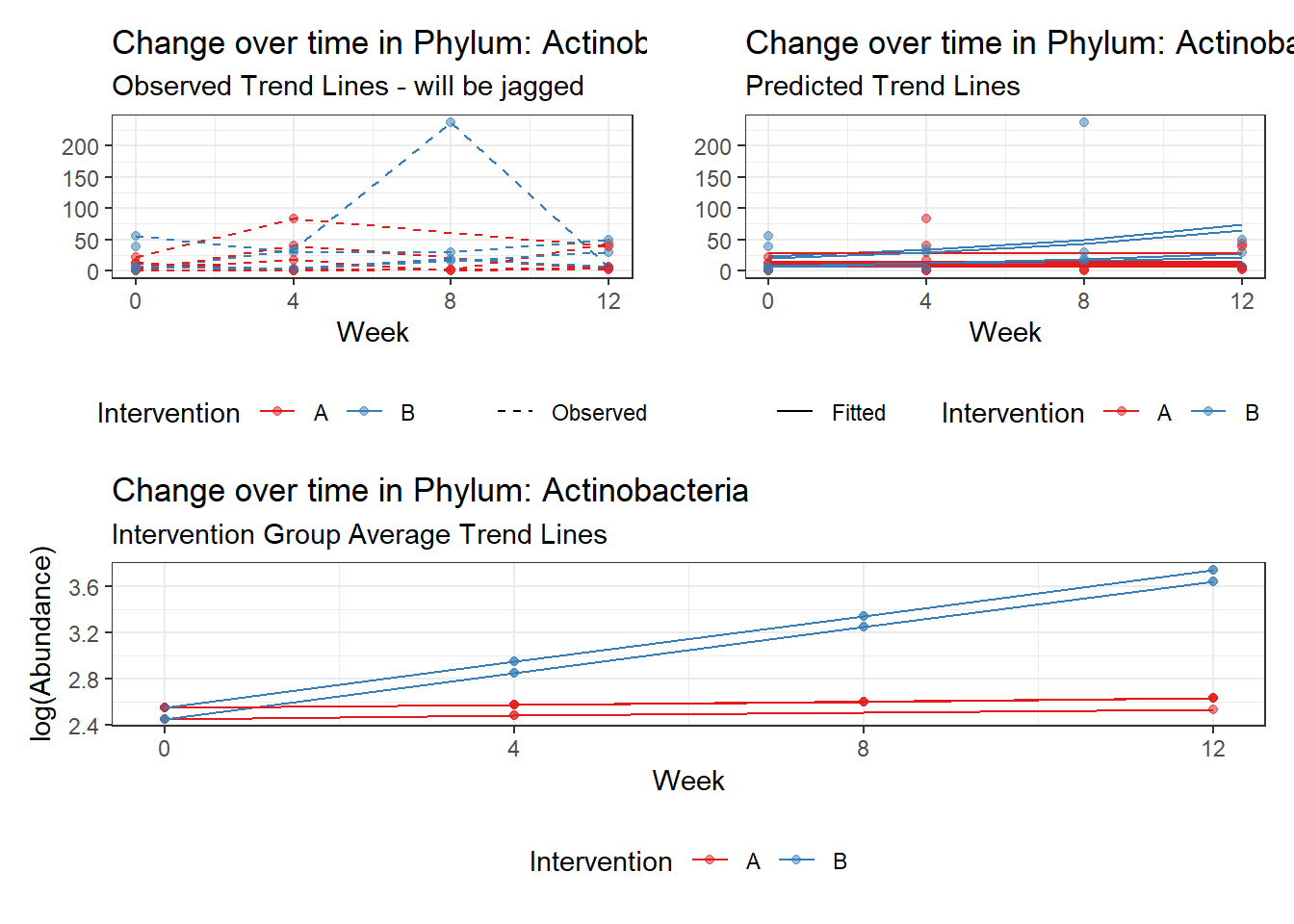
Covariate(s): Time and Ethnicity
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Ethnicity)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
phylum.fit8 <- glmm_microbiome(mydata=microbiome_data, model.number=8,
taxa.level="Phylum", link="poisson",
model="1 + time + intB:time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.4638933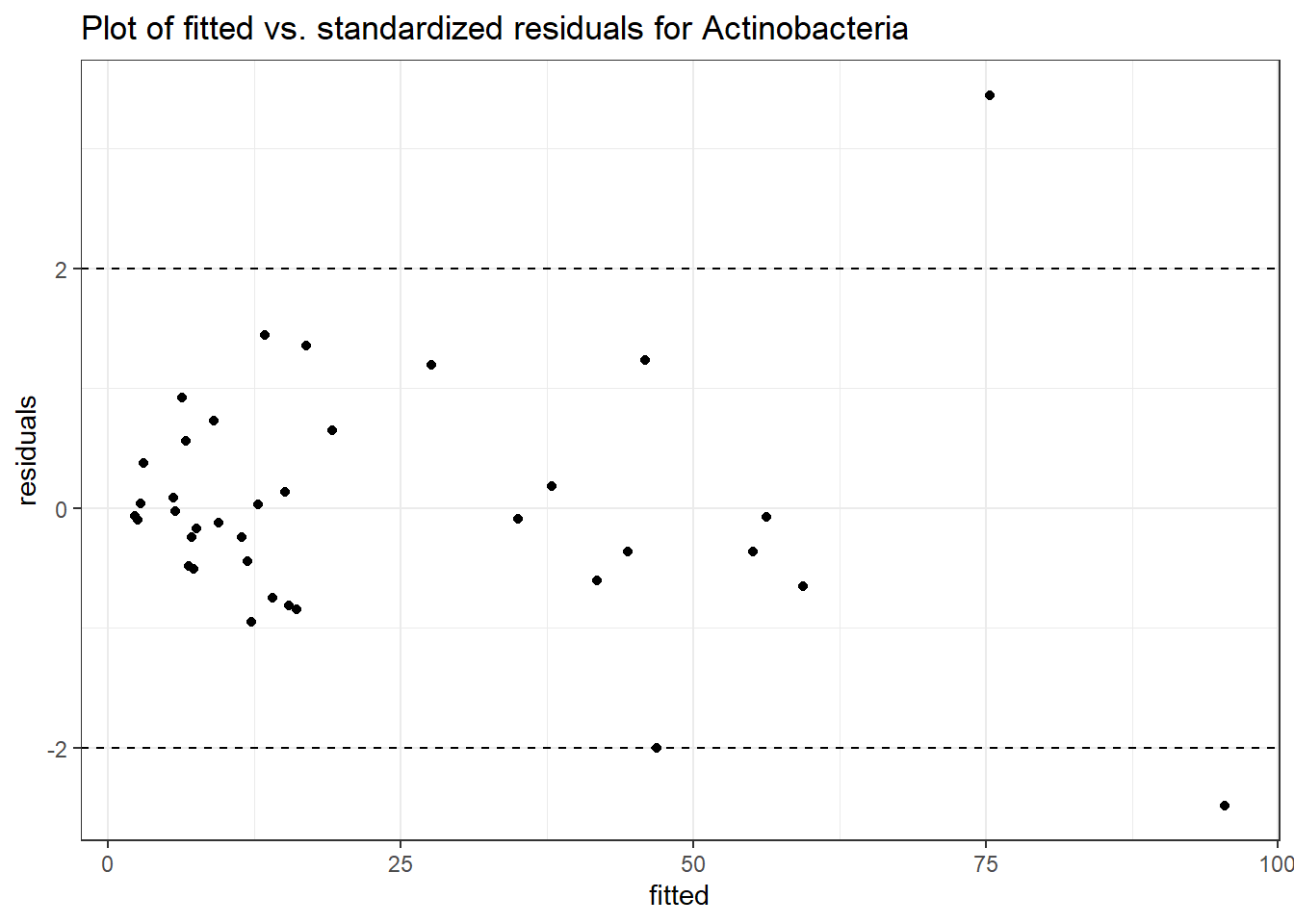
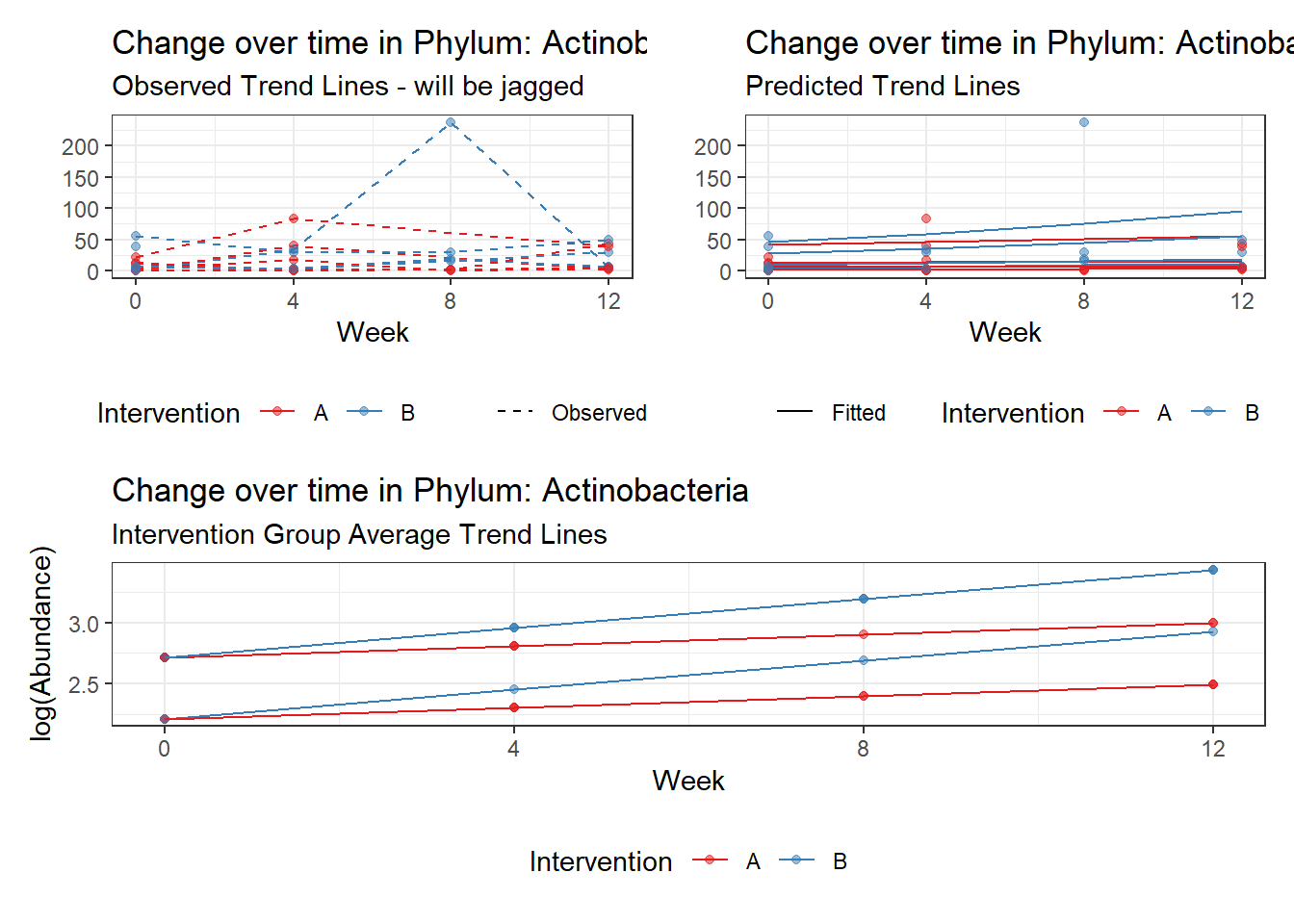
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.21500 0.47943 4.62002 0.00000
time Actinobacteria 0.09227 0.04834 1.90855 0.05632
hispanic Actinobacteria 0.50204 0.59533 0.84330 0.39906
time:intB Actinobacteria 0.14467 0.06212 2.32877 0.01987
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.93022
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.1637301

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.22001 0.22132 37.14023 0.00000
time Bacteroidetes -0.02184 0.00363 -6.01613 0.00000
hispanic Bacteroidetes -0.33195 0.27745 -1.19644 0.23153
time:intB Bacteroidetes -0.02242 0.00531 -4.22000 0.00002
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.44248
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.9381629

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -2.48209 2.47550 -1.00266 0.31602
time Cyanobacteria -0.07468 0.04113 -1.81560 0.06943
hispanic Cyanobacteria 2.78475 2.80609 0.99239 0.32101
time:intB Cyanobacteria -0.49553 0.06783 -7.30498 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.8951
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.5990631
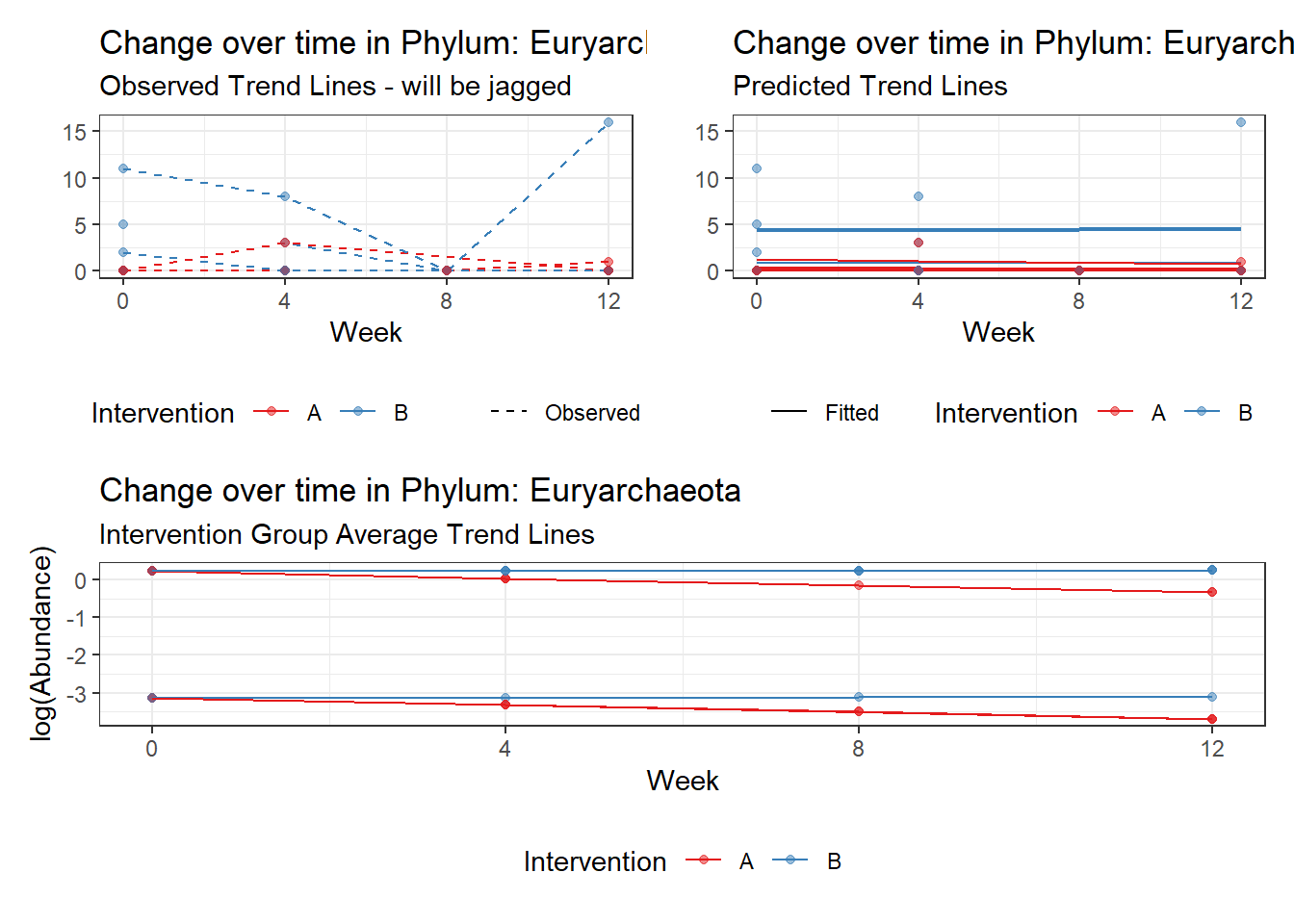
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -3.14003 1.39179 -2.25611 0.02406
time Euryarchaeota -0.18300 0.38038 -0.48110 0.63045
hispanic Euryarchaeota 3.36362 1.38916 2.42134 0.01546
time:intB Euryarchaeota 0.19488 0.40370 0.48275 0.62928
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.2224
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.1311736
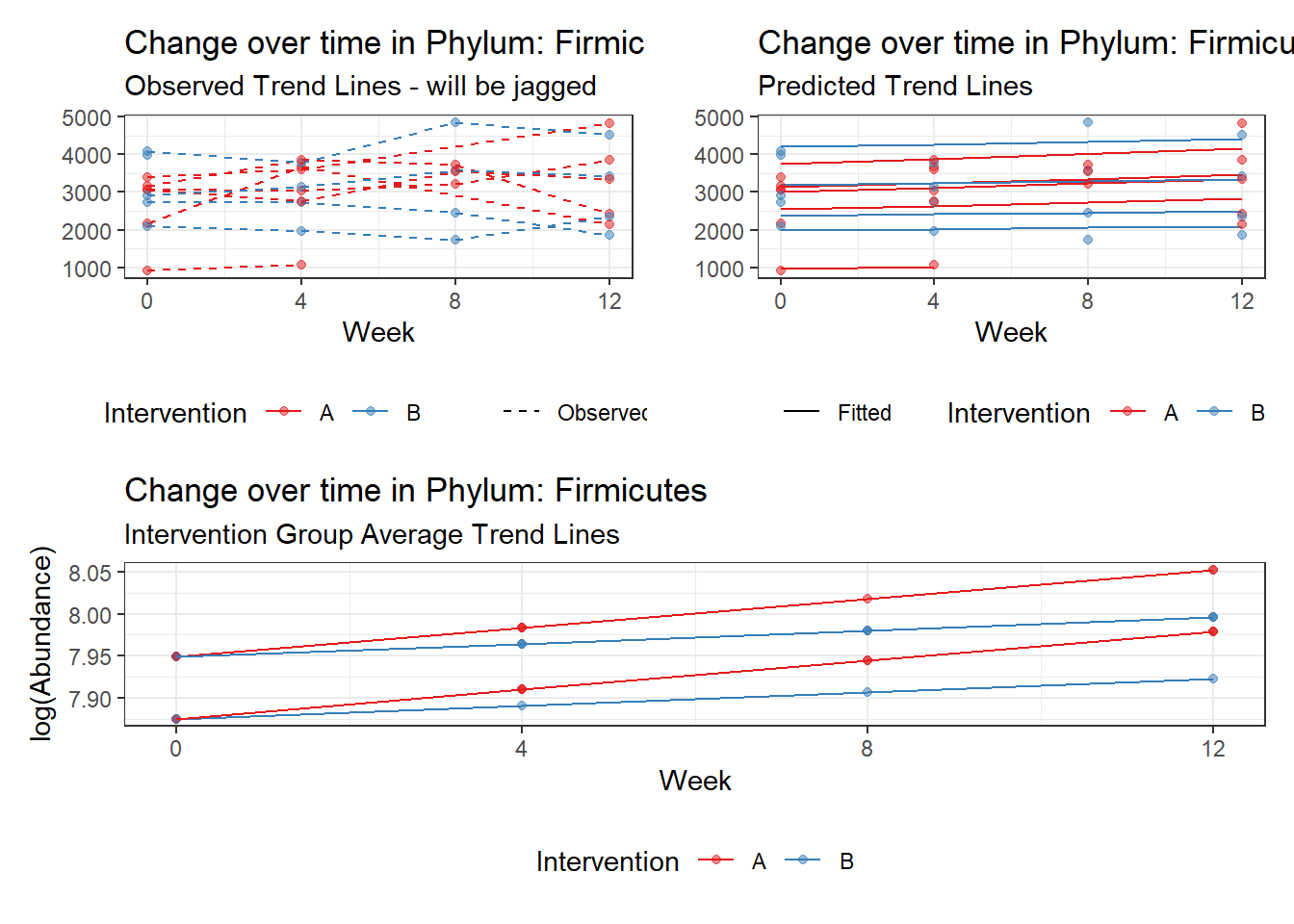
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.87502 0.19439 40.51150 0.00000
time Firmicutes 0.03458 0.00352 9.82039 0.00000
hispanic Firmicutes 0.07358 0.24366 0.30197 0.76268
time:intB Firmicutes -0.01873 0.00538 -3.47835 0.00050
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.38856
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.8737105
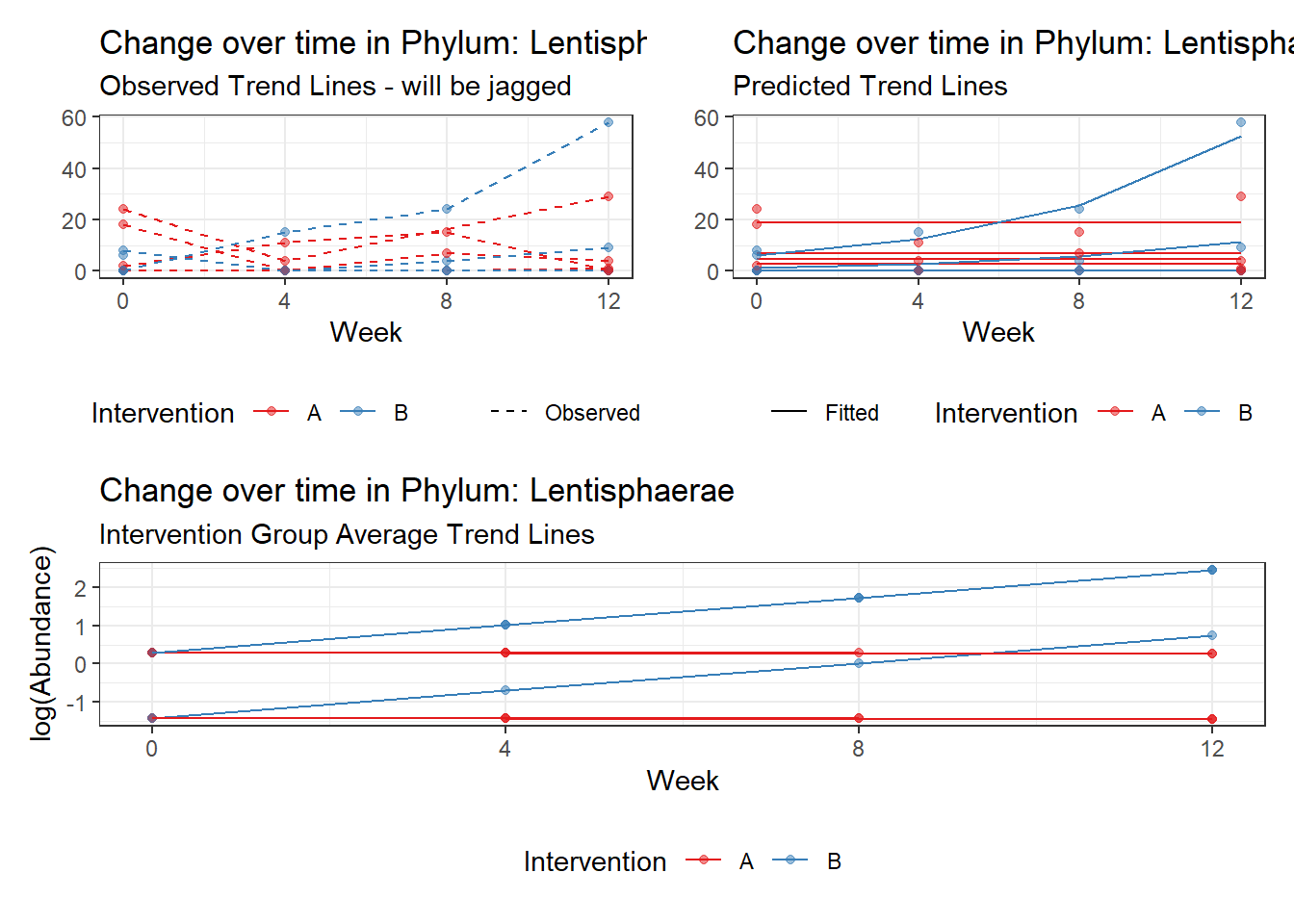
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae -1.41986 1.58979 -0.89311 0.37180
time Lentisphaerae -0.00384 0.07911 -0.04859 0.96125
hispanic Lentisphaerae 1.70539 1.84349 0.92509 0.35492
time:intB Lentisphaerae 0.72269 0.12740 5.67237 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6303
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.08999382
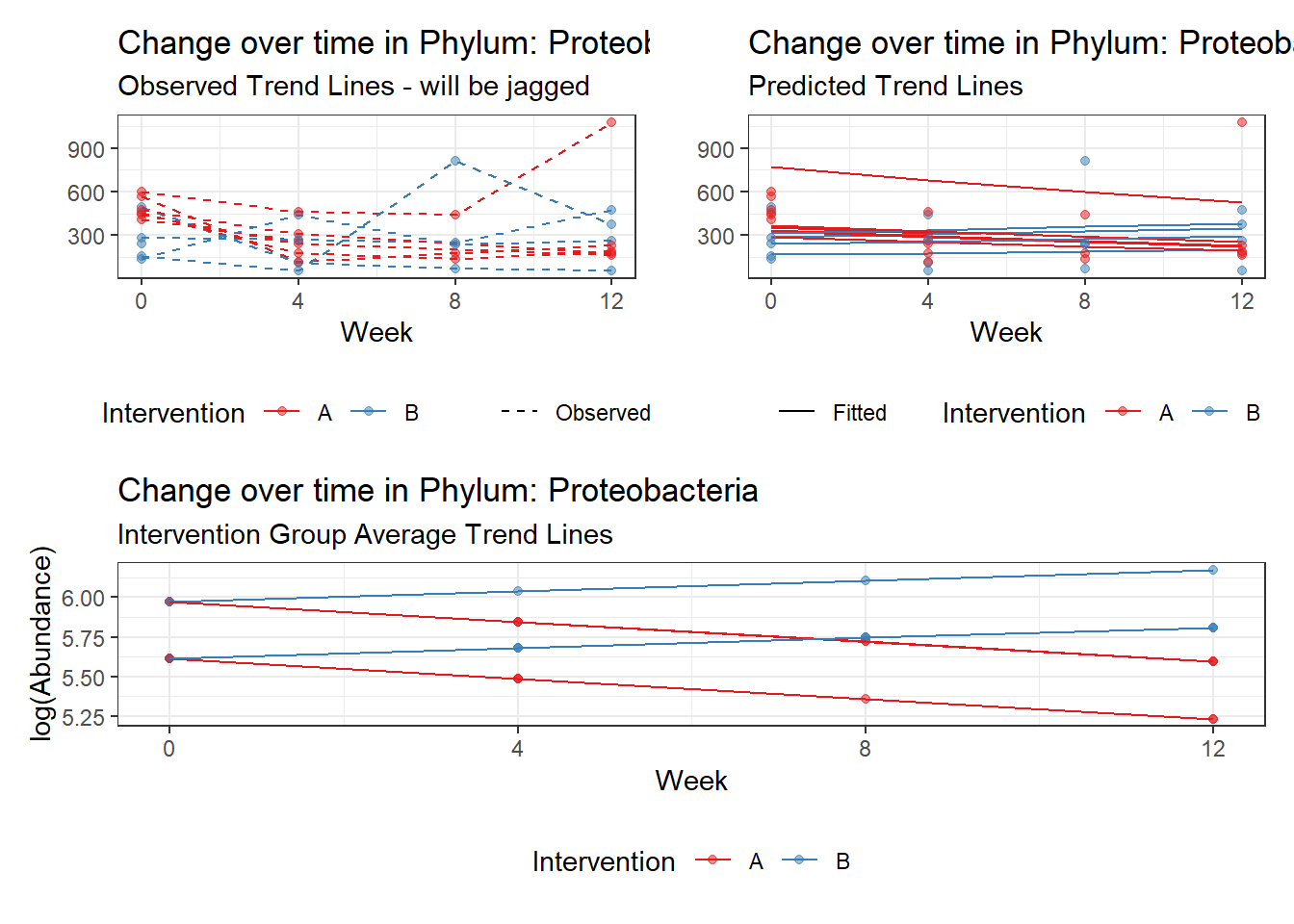
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 5.97256 0.15832 37.72526 0.00000
time Proteobacteria -0.12599 0.01087 -11.59167 0.00000
hispanic Proteobacteria -0.36129 0.19829 -1.82199 0.06846
time:intB Proteobacteria 0.19257 0.01721 11.18746 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.31447
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.9056279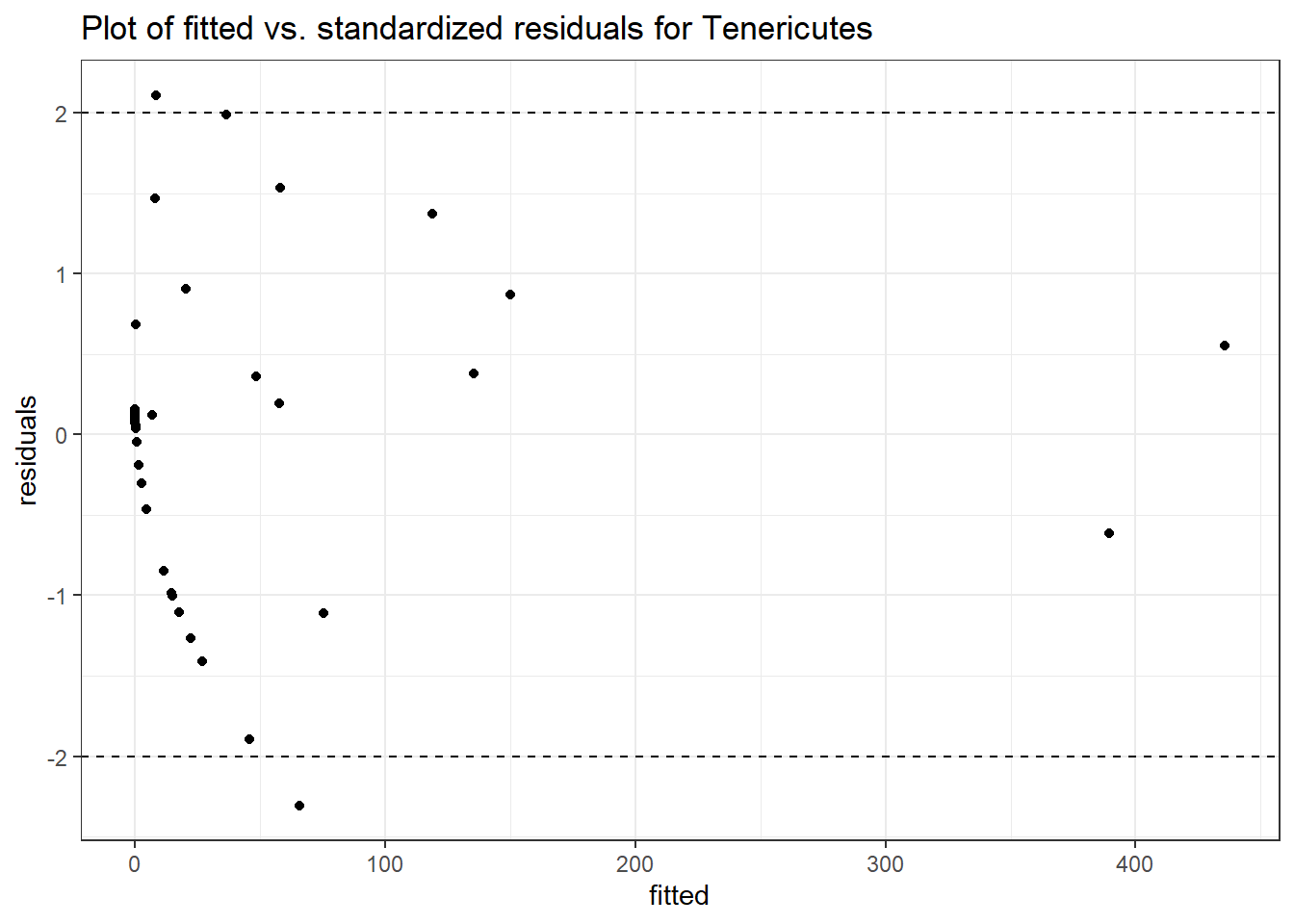

Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes -1.82957 1.82980 -0.99987 0.31737
time Tenericutes 0.58418 0.03029 19.28556 0.00000
hispanic Tenericutes 3.30359 2.15739 1.53129 0.12570
time:intB Tenericutes 0.36911 0.05297 6.96874 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.0978
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000004dc96ee0>
Link: poisson
Intraclass Correlation (ICC): 0.8527204
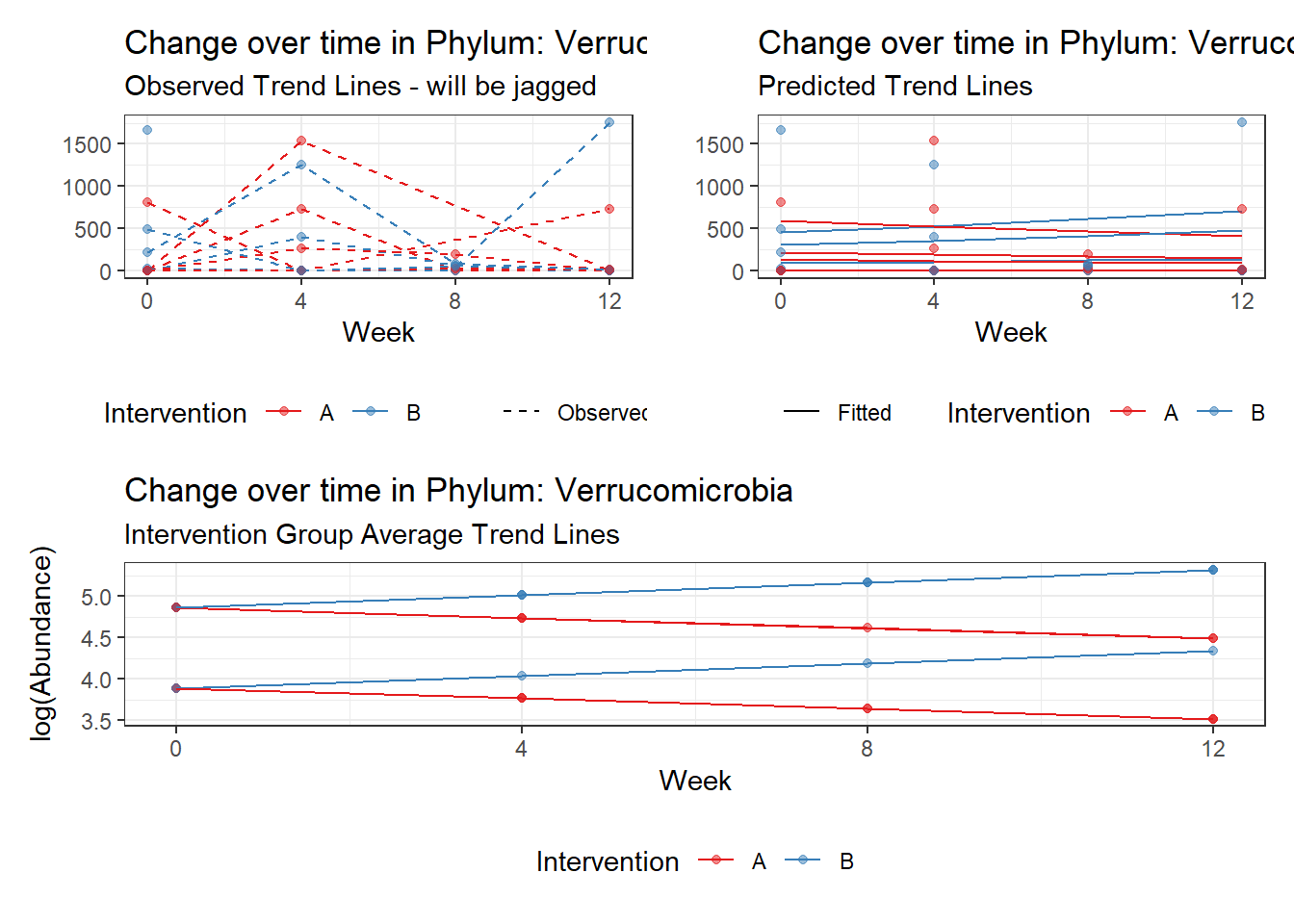
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 3.89165 1.20479 3.23014 0.00124
time Verrucomicrobia -0.12314 0.01296 -9.49942 0.00000
hispanic Verrucomicrobia 0.97273 1.51647 0.64145 0.52123
time:intB Verrucomicrobia 0.27345 0.01882 14.52637 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4062
Significance test not available for random effectsphylum.fit8b <- glmm_microbiome(mydata=microbiome_data, model.number=8,
taxa.level="Phylum", link="negbinom",
model="1 + time + intB:time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinom
Intraclass Correlation (ICC): 0.3139274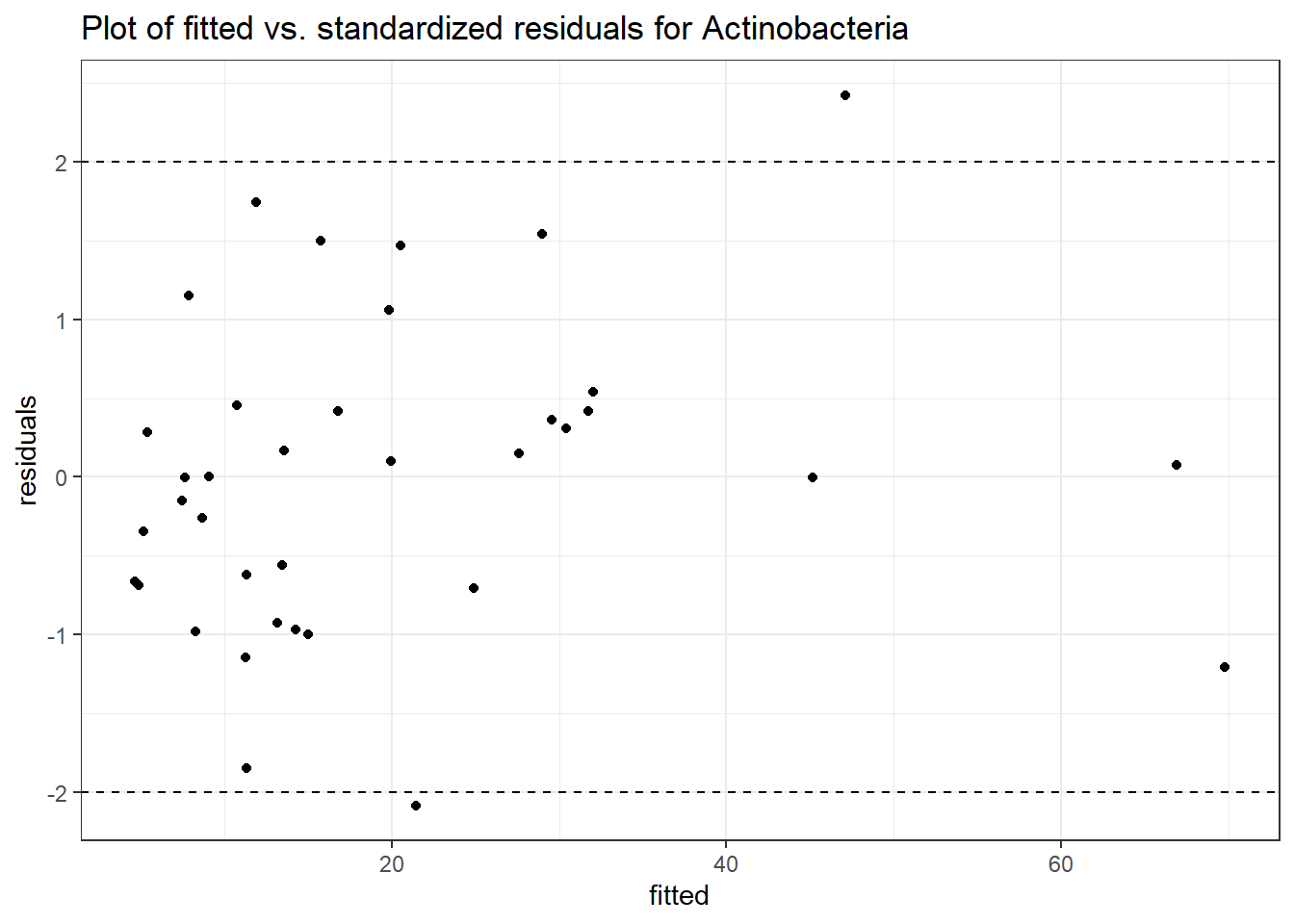
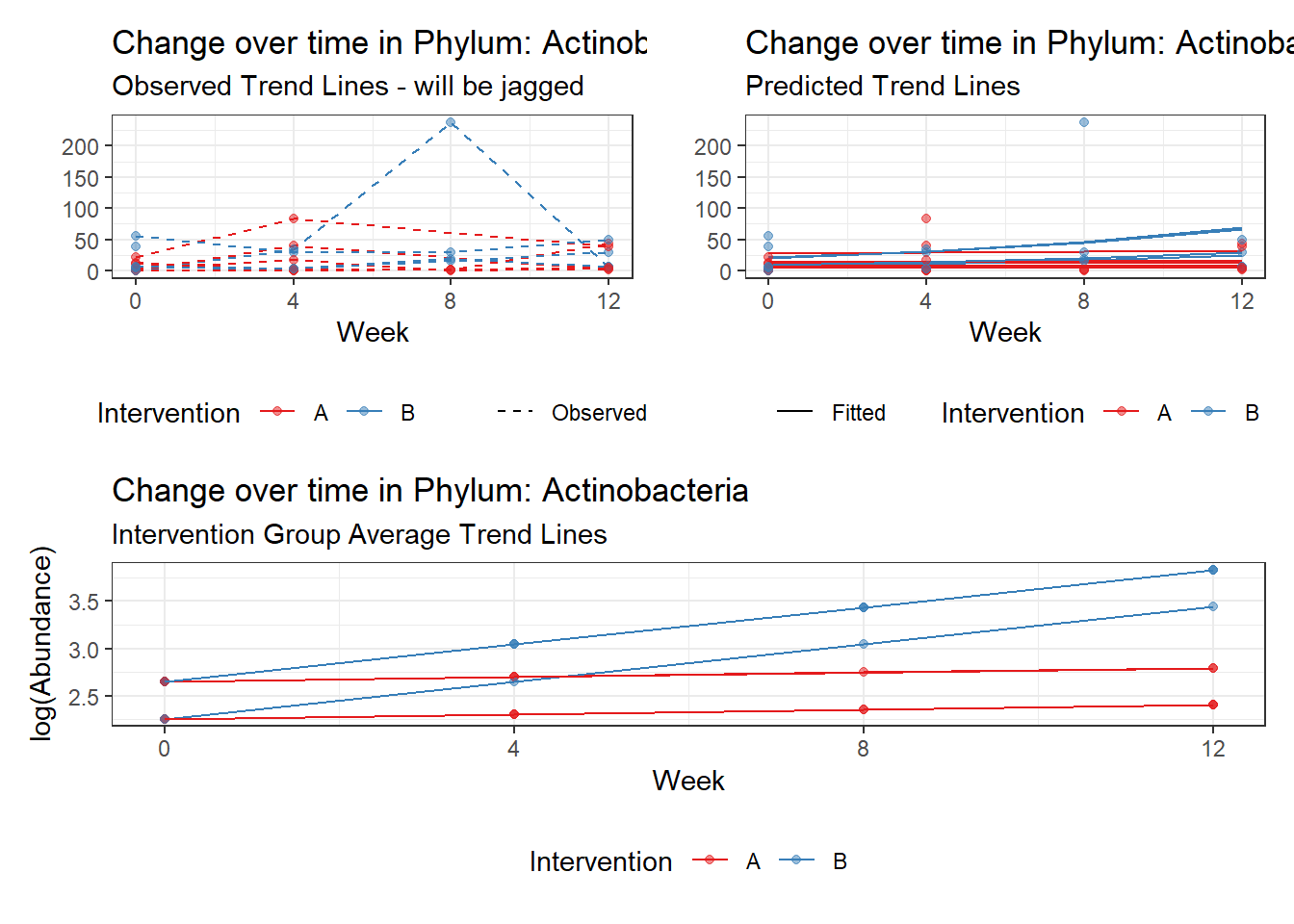
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.25734 0.55848 4.04197 0.00005
time Actinobacteria 0.04951 0.22993 0.21534 0.82950
hispanic Actinobacteria 0.39075 0.59336 0.65853 0.51020
time:intB Actinobacteria 0.34387 0.28856 1.19165 0.23340
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.67644
Significance test not available for random effects
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinom
Intraclass Correlation (ICC): 0.125987
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.24721 0.20728 39.78687 0.00000
time Bacteroidetes -0.03888 0.04534 -0.85753 0.39115
hispanic Bacteroidetes -0.31467 0.25474 -1.23523 0.21674
time:intB Bacteroidetes -0.02152 0.06644 -0.32388 0.74603
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.37967
Significance test not available for random effects
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinomWarning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
iteration limit reachedWarning in sqrt(1/i): NaNs produced
Intraclass Correlation (ICC): 0.9386044
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria -2.51414 2.49472 -1.00778 0.31356
time Cyanobacteria -0.07242 0.04207 -1.72128 0.08520
hispanic Cyanobacteria 2.80768 2.76933 1.01385 0.31066
time:intB Cyanobacteria -0.50406 0.06895 -7.31046 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.91
Significance test not available for random effects
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 4.924077e-12
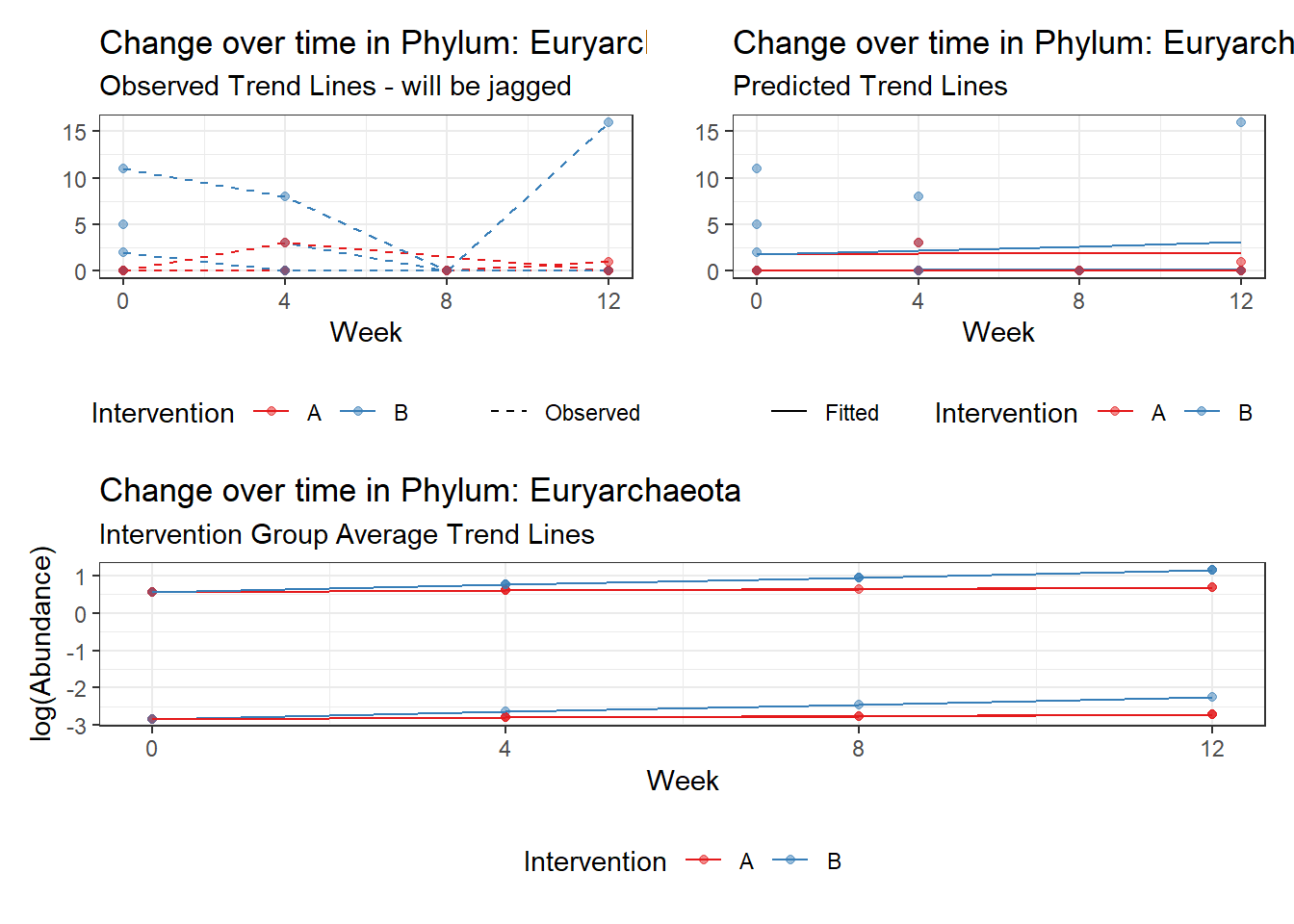
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -2.83063 1.78173 -1.58870 0.11213
time Euryarchaeota 0.03947 0.78617 0.05020 0.95996
hispanic Euryarchaeota 3.40024 1.75962 1.93237 0.05331
time:intB Euryarchaeota 0.15439 0.81138 0.19028 0.84909
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.219e-06
Significance test not available for random effects
#########################################
#########################################
Model: Firmicutes ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinom
Intraclass Correlation (ICC): 0.1193617
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 7.88045 0.19209 41.02414 0.00000
time Firmicutes 0.03450 0.03213 1.07361 0.28300
hispanic Firmicutes 0.07388 0.23822 0.31015 0.75644
time:intB Firmicutes -0.02733 0.04668 -0.58553 0.55819
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.36816
Significance test not available for random effects
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinomboundary (singular) fit: see ?isSingular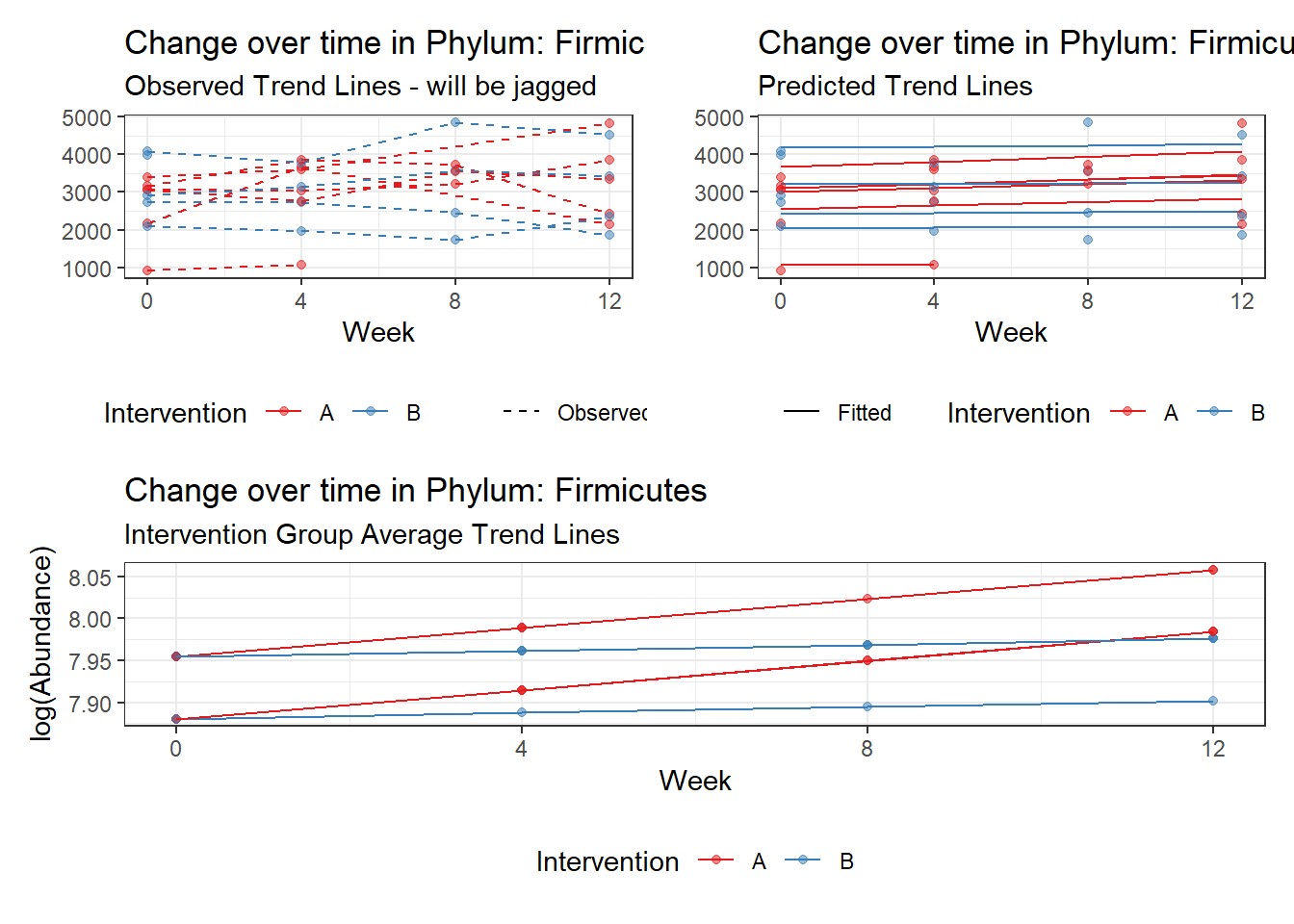
Intraclass Correlation (ICC): 1.405955e-11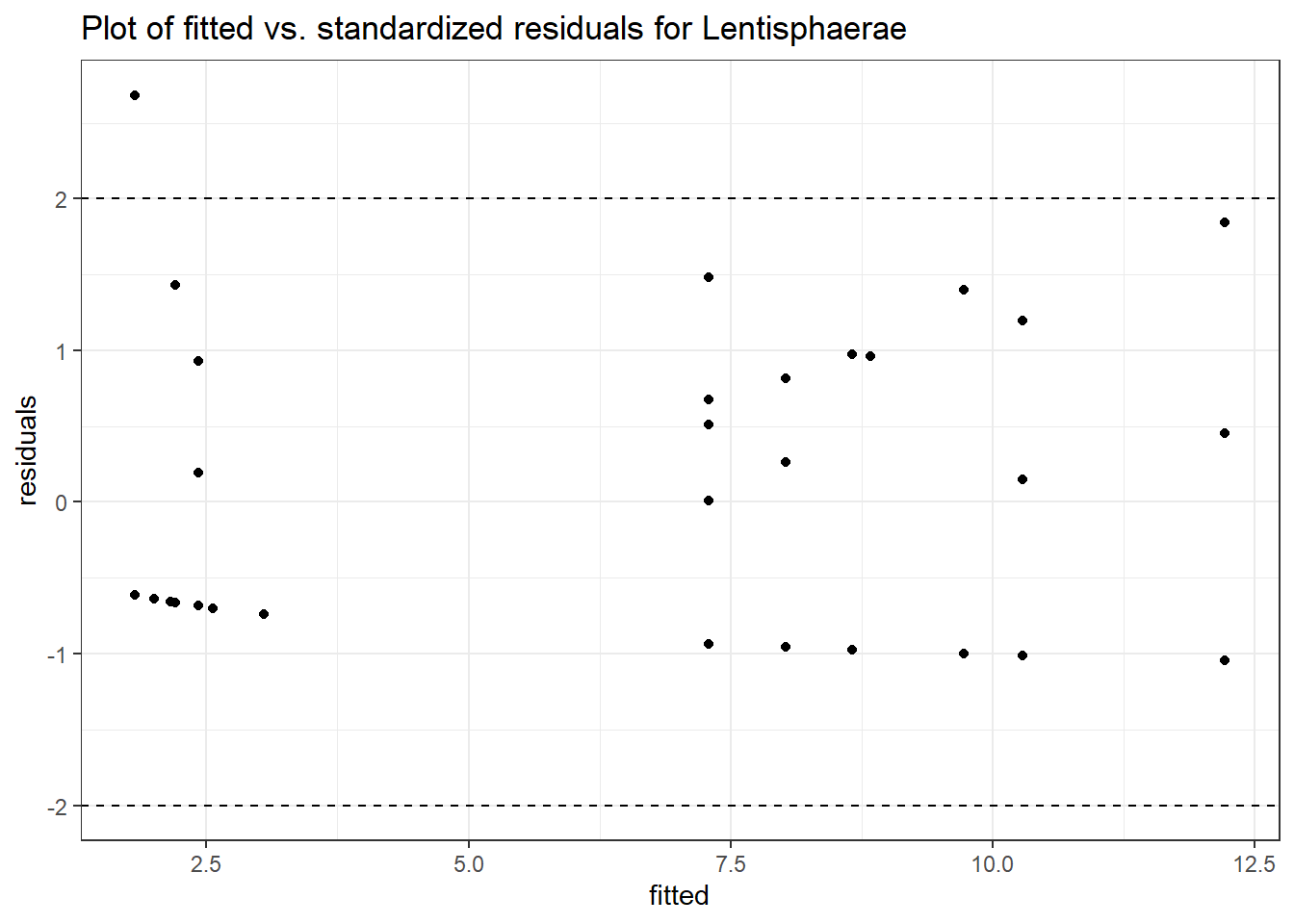
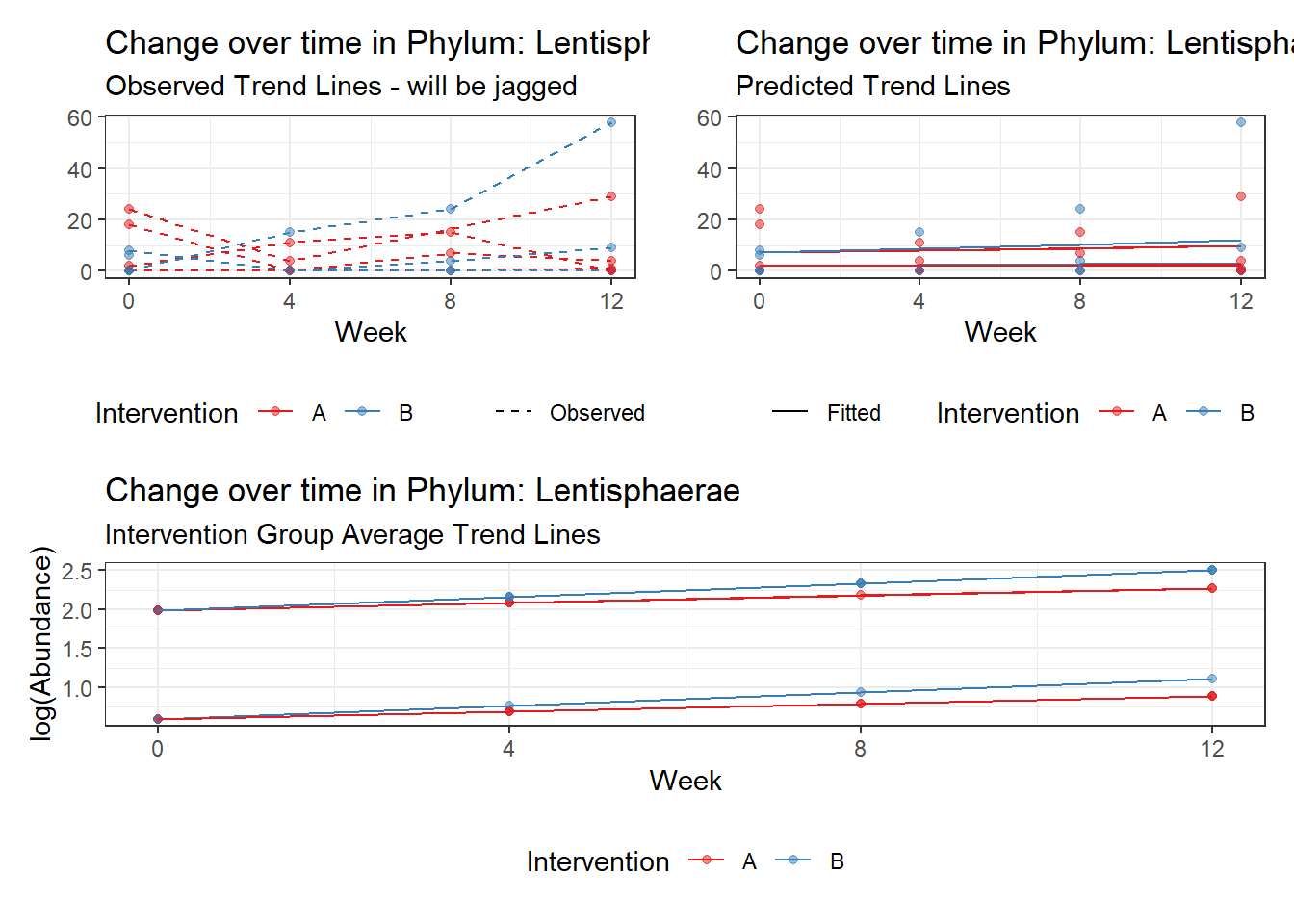
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.59857 0.65048 0.92019 0.35747
time Lentisphaerae 0.09582 0.34241 0.27983 0.77961
hispanic Lentisphaerae 1.38859 0.82312 1.68699 0.09161
time:intB Lentisphaerae 0.07602 0.40262 0.18882 0.85023
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.7496e-06
Significance test not available for random effects
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinom
Intraclass Correlation (ICC): 0.01340966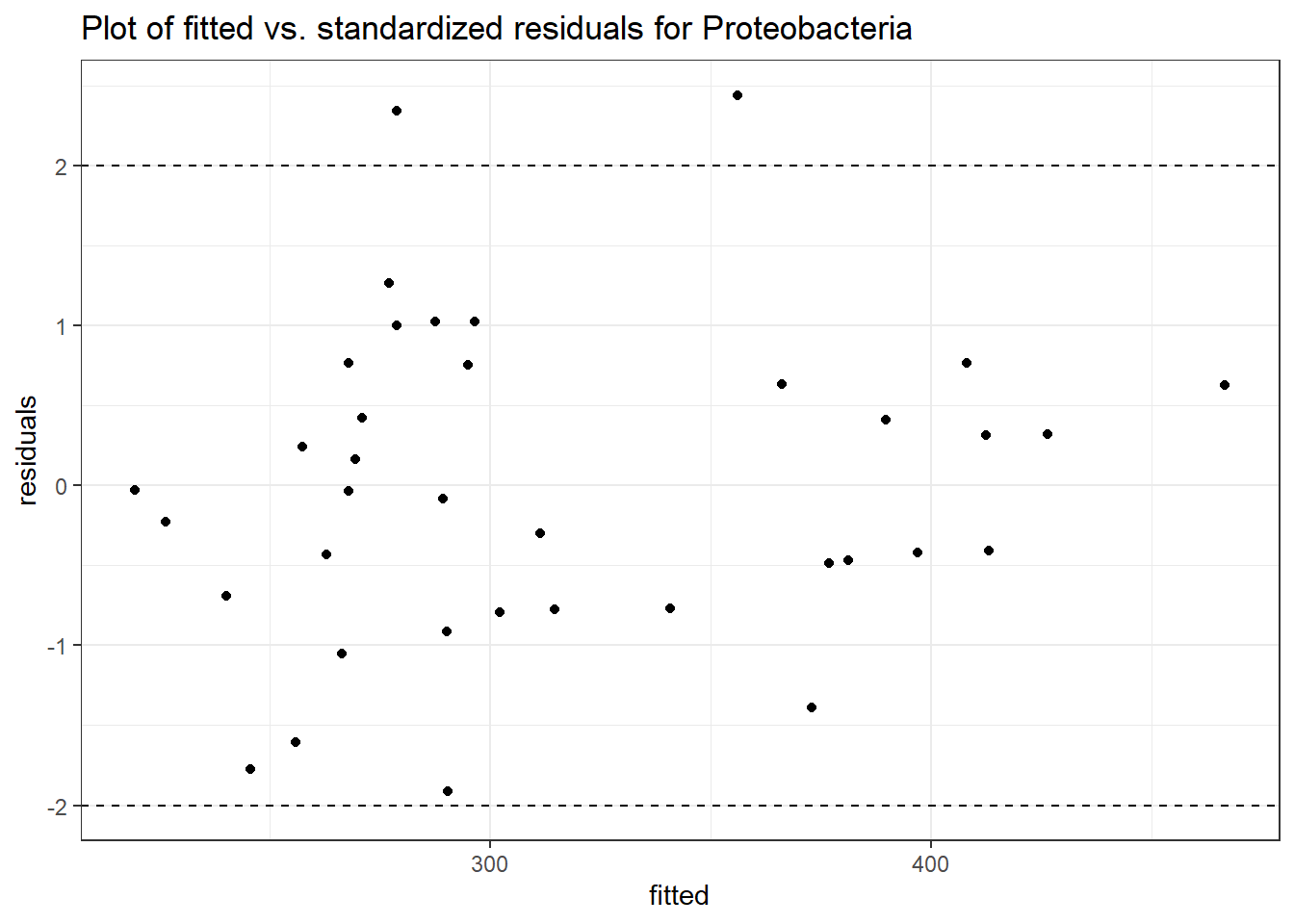
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.05066 0.21812 27.73948 0.00000
time Proteobacteria -0.09016 0.10934 -0.82461 0.40960
hispanic Proteobacteria -0.37745 0.23041 -1.63818 0.10138
time:intB Proteobacteria 0.04997 0.12449 0.40141 0.68811
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.11658
Significance test not available for random effects
#########################################
#########################################
Model: Tenericutes ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinomboundary (singular) fit: see ?isSingular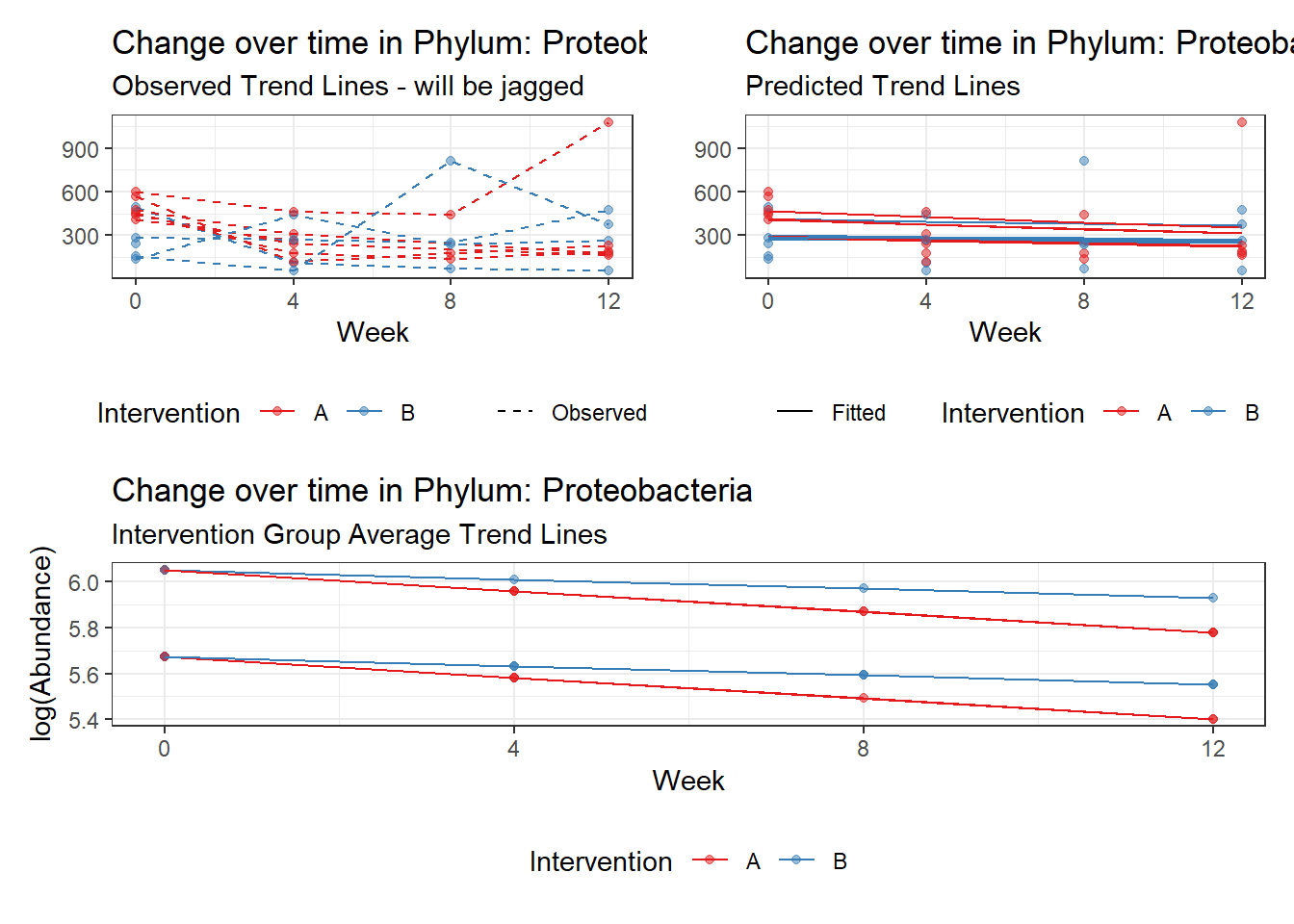
Intraclass Correlation (ICC): 1.936714e-11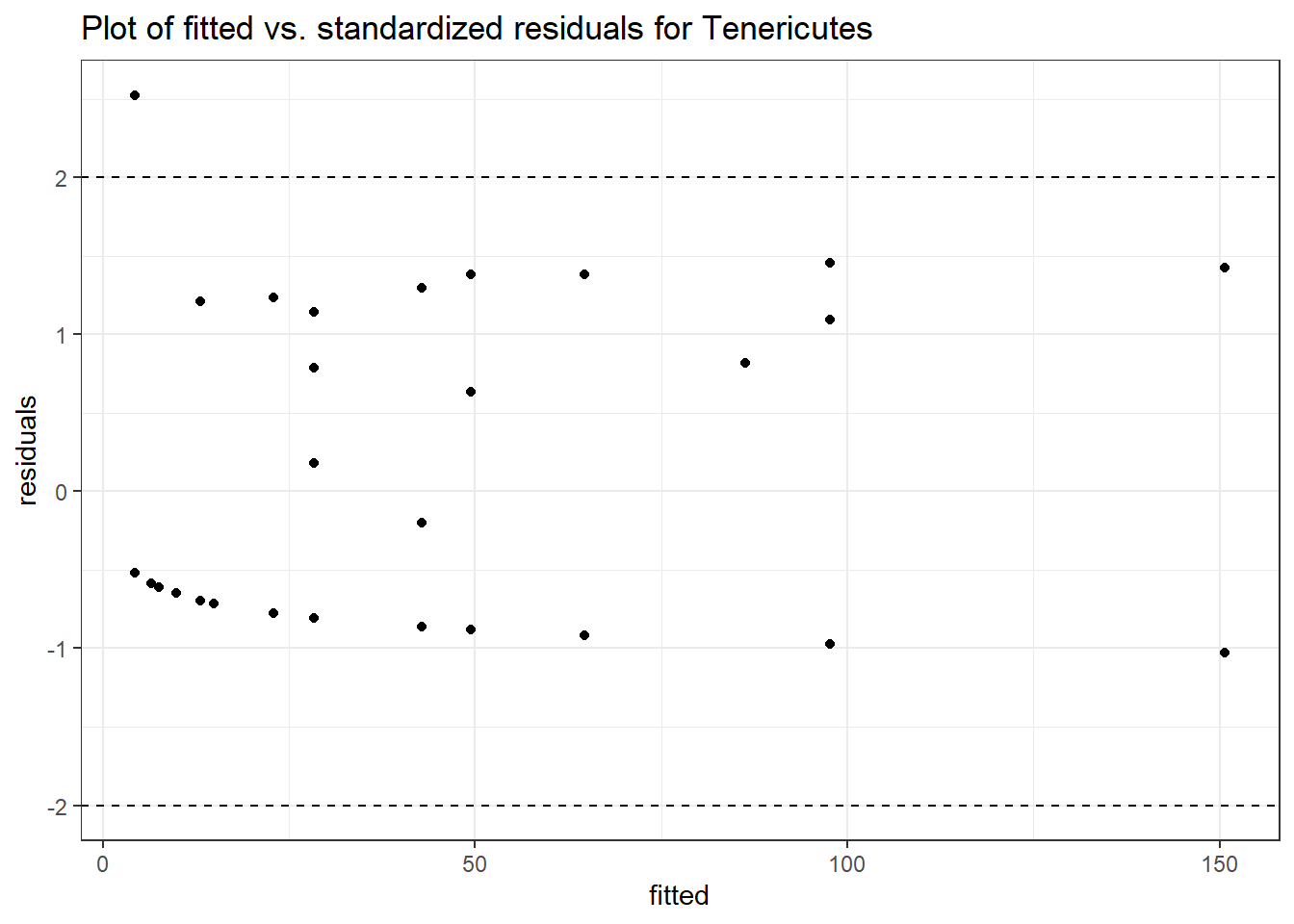
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 1.46073 0.86963 1.67972 0.09301
time Tenericutes 0.55723 0.48228 1.15541 0.24792
hispanic Tenericutes 1.88305 1.16936 1.61033 0.10733
time:intB Tenericutes -0.14473 0.56702 -0.25526 0.79853
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 4.4008e-06
Significance test not available for random effects
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + intB:time + hispanic + (1 | SubjectID)
<environment: 0x000000000b990df8>
Link: negbinomboundary (singular) fit: see ?isSingular
Intraclass Correlation (ICC): 4.148378e-12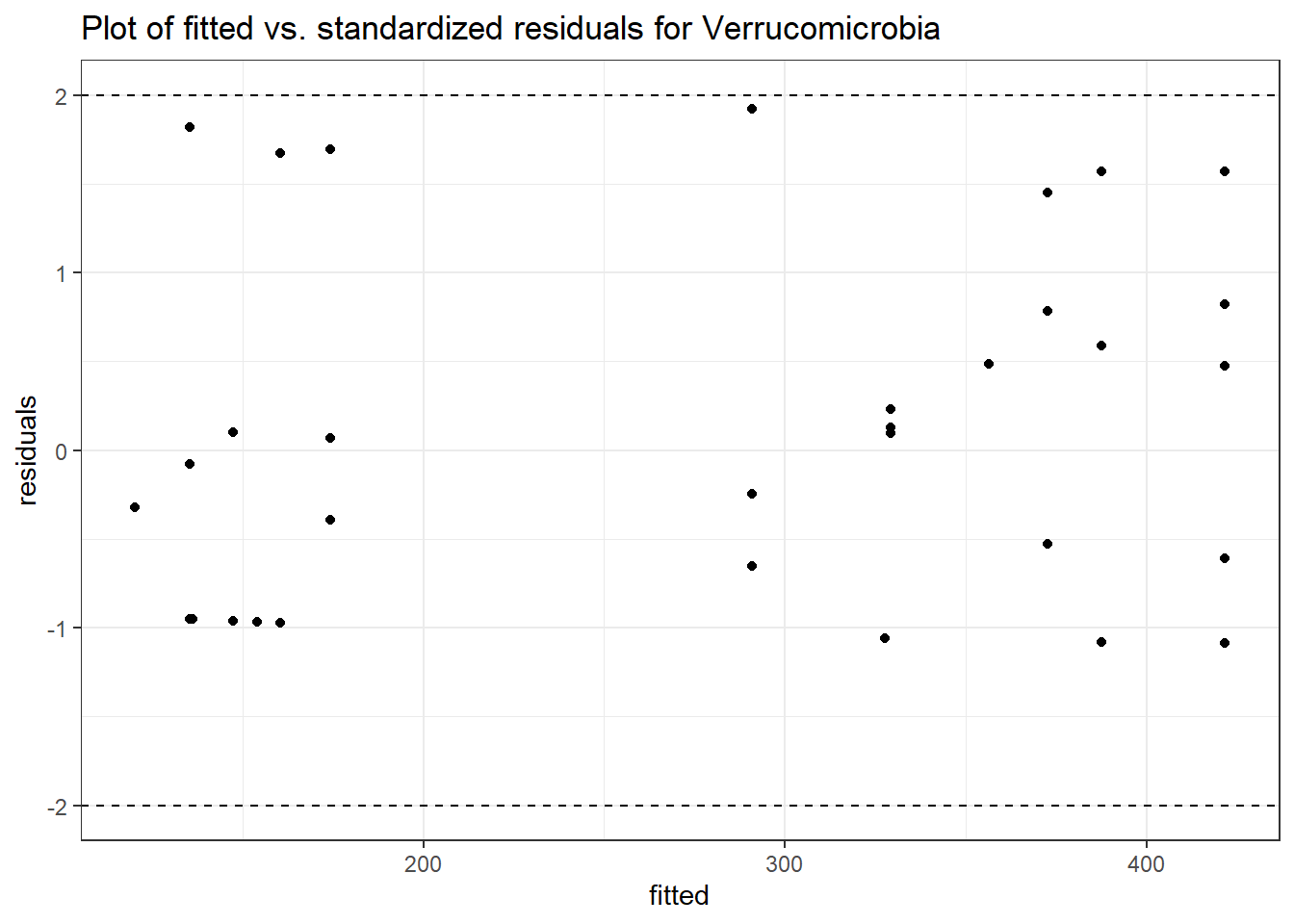
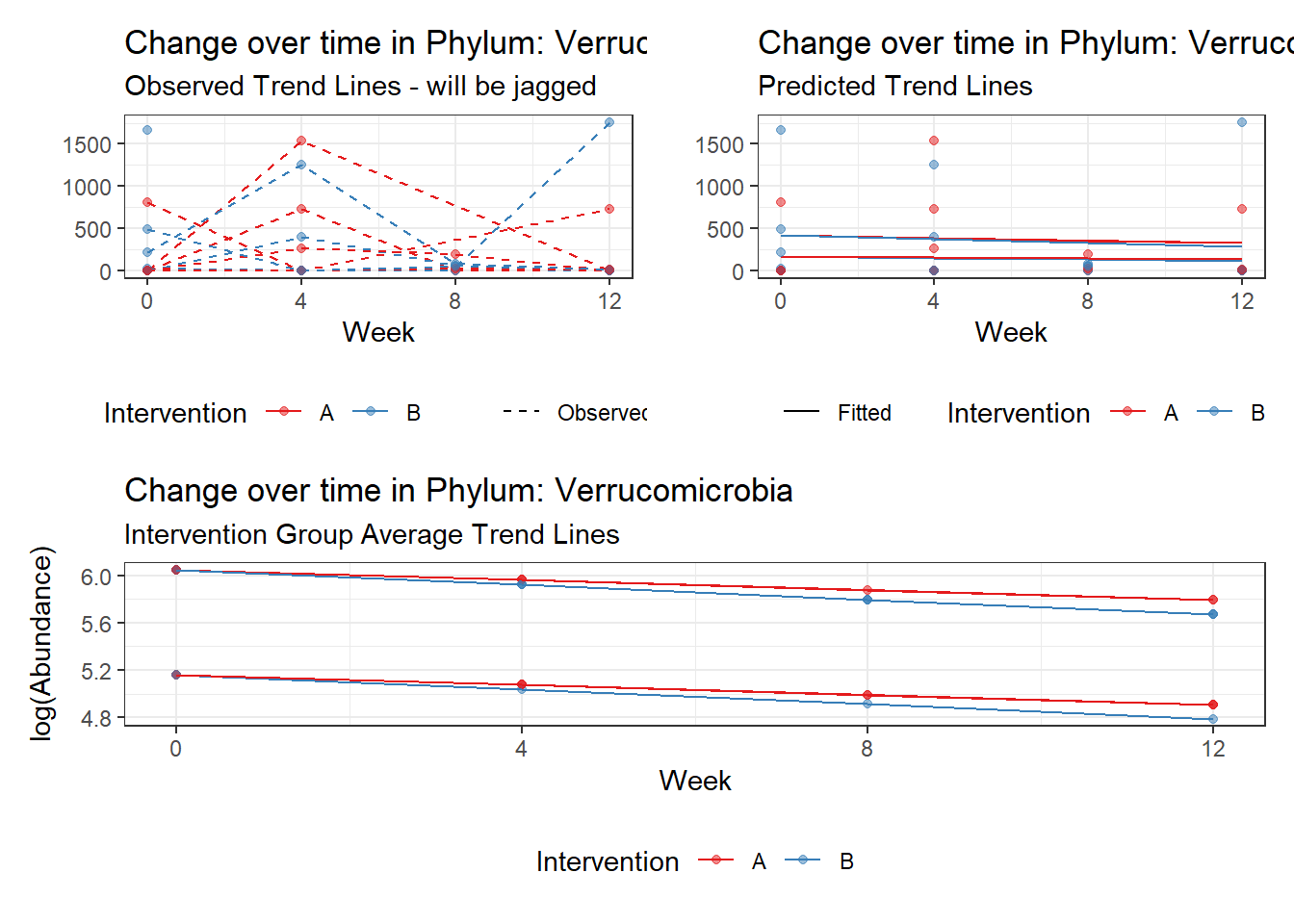
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 5.16109 0.93773 5.50382 0.00000
time Verrucomicrobia -0.08415 0.47434 -0.17739 0.85920
hispanic Verrucomicrobia 0.88331 1.06565 0.82889 0.40717
time:intB Verrucomicrobia -0.03976 0.57503 -0.06914 0.94488
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.0368e-06
Significance test not available for random effectsSummary of Effect of Intervention on Abundance
To summarize these results, we are looking 1 main things: that the interaction between intervention and time (intB:time) is significant. If so, then we have evidence that that phylum was significantly affected by the intervention, meaning that as time progresses those in the intervention had their microbiome abundance differentially changed over time.
This is sumarized in two ways (i.e., two models). These models are detailed below.
Model 6: Intervention by Time Interaction Only
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\] where, the major aim was to assess the significance of two terms:
- the interaction between time and intervention (intB:time) (\(\gamma_{11}\)) - which assessing the magnitude of the change in alpha diversity as the intervention progresses.
model6.results <- unlist_results(phylum.fit6b$`Fixed Effects`)Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")kable(model6.results, format="html", digits=3)%>%
kable_styling(full_width = T) %>%
scroll_box(width="100%",height="5in")| Phylum | Term | Estimate | Std. Error | z value | Pr(>|z|) | p.adjust |
|---|---|---|---|---|---|---|
| Actinobacteria | time | 0.033 | 0.227 | 0.144 | 0.885 | 0.952 |
| Actinobacteria | time:intB | 0.356 | 0.285 | 1.250 | 0.211 | 0.475 |
| Bacteroidetes | time | -0.033 | 0.045 | -0.724 | 0.469 | 0.777 |
| Bacteroidetes | time:intB | -0.032 | 0.066 | -0.486 | 0.627 | 0.777 |
| Cyanobacteria | time | -0.073 | 0.042 | -1.725 | 0.085 | 0.275 |
| Cyanobacteria | time:intB | -0.504 | 0.069 | -7.302 | 0.000 | 0.000 |
| Euryarchaeota | time | -0.836 | 0.574 | -1.456 | 0.145 | 0.392 |
| Euryarchaeota | time:intB | 1.043 | 0.618 | 1.687 | 0.092 | 0.275 |
| Firmicutes | time | 0.034 | 0.032 | 1.054 | 0.292 | 0.606 |
| Firmicutes | time:intB | -0.026 | 0.047 | -0.564 | 0.573 | 0.777 |
| Lentisphaerae | time | 0.033 | 0.363 | 0.091 | 0.928 | 0.952 |
| Lentisphaerae | time:intB | 0.243 | 0.509 | 0.477 | 0.633 | 0.777 |
| Proteobacteria | time | -0.072 | 0.110 | -0.657 | 0.511 | 0.777 |
| Proteobacteria | time:intB | 0.018 | 0.131 | 0.134 | 0.893 | 0.952 |
| Tenericutes | time | 0.691 | 0.533 | 1.295 | 0.195 | 0.475 |
| Tenericutes | time:intB | 0.035 | 0.579 | 0.060 | 0.952 | 0.952 |
| Verrucomicrobia | time | -0.276 | 0.447 | -0.619 | 0.536 | 0.777 |
| Verrucomicrobia | time:intB | 0.236 | 0.468 | 0.504 | 0.614 | 0.777 |
Dual results for double check
model6.pois.nb.results <- unlist_dual_results(phylum.fit6$`Fixed Effects`, phylum.fit6b$`Fixed Effects`)Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")
Joining, by = c("Phylum", "Term", "Estimate", "Std. Error", "z value", "Pr(>|z|)")kable(model6.pois.nb.results, format="html", digits=3)%>%
kable_styling(full_width = T) %>%
add_header_above(c('Poisson Model'=7, 'Negative Binomial Model'=7)) %>%
scroll_box(width="100%",height="5in")| pois.Phylum | pois.Term | pois.Estimate | pois.Std. Error | pois.z value | pois.Pr(>|z|) | pois.p.adjust | nb.Phylum | nb.Term | nb.Estimate | nb.Std. Error | nb.z value | nb.Pr(>|z|) | nb.p.adjust |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Actinobacteria | time | 0.091 | 0.048 | 1.874 | 0.061 | 0.087 | Actinobacteria | time | 0.033 | 0.227 | 0.144 | 0.885 | 0.952 |
| Actinobacteria | time:intB | 0.147 | 0.062 | 2.365 | 0.018 | 0.027 | Actinobacteria | time:intB | 0.356 | 0.285 | 1.250 | 0.211 | 0.475 |
| Bacteroidetes | time | -0.022 | 0.004 | -6.008 | 0.000 | 0.000 | Bacteroidetes | time | -0.033 | 0.045 | -0.724 | 0.469 | 0.777 |
| Bacteroidetes | time:intB | -0.022 | 0.005 | -4.230 | 0.000 | 0.000 | Bacteroidetes | time:intB | -0.032 | 0.066 | -0.486 | 0.627 | 0.777 |
| Cyanobacteria | time | -0.075 | 0.041 | -1.826 | 0.068 | 0.092 | Cyanobacteria | time | -0.073 | 0.042 | -1.725 | 0.085 | 0.275 |
| Cyanobacteria | time:intB | -0.495 | 0.068 | -7.297 | 0.000 | 0.000 | Cyanobacteria | time:intB | -0.504 | 0.069 | -7.302 | 0.000 | 0.000 |
| Euryarchaeota | time | -0.194 | 0.385 | -0.503 | 0.615 | 0.701 | Euryarchaeota | time | -0.836 | 0.574 | -1.456 | 0.145 | 0.392 |
| Euryarchaeota | time:intB | 0.201 | 0.410 | 0.491 | 0.623 | 0.701 | Euryarchaeota | time:intB | 1.043 | 0.618 | 1.687 | 0.092 | 0.275 |
| Firmicutes | time | 0.035 | 0.004 | 9.818 | 0.000 | 0.000 | Firmicutes | time | 0.034 | 0.032 | 1.054 | 0.292 | 0.606 |
| Firmicutes | time:intB | -0.019 | 0.005 | -3.476 | 0.001 | 0.001 | Firmicutes | time:intB | -0.026 | 0.047 | -0.564 | 0.573 | 0.777 |
| Lentisphaerae | time | -0.006 | 0.079 | -0.082 | 0.935 | 0.935 | Lentisphaerae | time | 0.033 | 0.363 | 0.091 | 0.928 | 0.952 |
| Lentisphaerae | time:intB | 0.729 | 0.127 | 5.732 | 0.000 | 0.000 | Lentisphaerae | time:intB | 0.243 | 0.509 | 0.477 | 0.633 | 0.777 |
| Proteobacteria | time | -0.126 | 0.011 | -11.569 | 0.000 | 0.000 | Proteobacteria | time | -0.072 | 0.110 | -0.657 | 0.511 | 0.777 |
| Proteobacteria | time:intB | 0.193 | 0.017 | 11.174 | 0.000 | 0.000 | Proteobacteria | time:intB | 0.018 | 0.131 | 0.134 | 0.893 | 0.952 |
| Tenericutes | time | 0.583 | 0.030 | 19.297 | 0.000 | 0.000 | Tenericutes | time | 0.691 | 0.533 | 1.295 | 0.195 | 0.475 |
| Tenericutes | time:intB | 0.371 | 0.053 | 7.015 | 0.000 | 0.000 | Tenericutes | time:intB | 0.035 | 0.579 | 0.060 | 0.952 | 0.952 |
| Verrucomicrobia | time | -0.123 | 0.013 | -9.503 | 0.000 | 0.000 | Verrucomicrobia | time | -0.276 | 0.447 | -0.619 | 0.536 | 0.777 |
| Verrucomicrobia | time:intB | 0.274 | 0.019 | 14.530 | 0.000 | 0.000 | Verrucomicrobia | time:intB | 0.236 | 0.468 | 0.504 | 0.614 | 0.777 |
write.csv(model6.pois.nb.results, "tab/results_glmm_microbiome_phylum.csv")Nice Plot
N <- nrow(microbiome_data$meta.dat)
ids <- microbiome_data$meta.dat$ID
dat <- data.frame(microbiome_data$abund.list[["Phylum"]])
# NOTE: Special coding for stripping weird characters from bacteria names
j <- length(rownames(dat))
i <- 1
for(i in 1:j){
while( substring(rownames(dat[i,]), 1, 1) == "_"){
if(i == 1){
row.names(dat) <- c(substring(rownames(dat[i,]), 2),rownames(dat)[2:j])
}
if(i > 1 & i < j){
row.names(dat) <- c(rownames(dat[1:(i-1),]),
substring(rownames(dat[i,]), 2),
rownames(dat)[(i+1):j])
}
if(i == j){
row.names(dat) <- c(rownames(dat[1:(j-1),]),substring(rownames(dat[j,]), 2))
}
} # End while loop
} # End for loop
# ====================== #
num.bact <- nrow(dat)
dat <- t(dat[1:num.bact,1:N])
dat <- apply(dat, 2, function(x){log(x+1)})
k <- ncol(microbiome_data$meta.dat) # number of original variables
dat <- data.frame(cbind(microbiome_data$meta.dat, dat[ids,]))
# first phylum
PH <- names(phylum.fit6b$`Fitted Models`)
pL <- list()
i <- 1
for(i in 1:length(PH)){
fit <- phylum.fit6b$`Fitted Models`[[PH[i]]]
idat <- cbind(dat, fit=exp(predict(fit)))
idat <- idat %>% mutate(Week = (as.numeric(Week)-1)*4)
idat$PRED <- predict(fit, re.form=NA)
idat$Outcome <- idat[, PH[i]]
# extract p-value
pv <- filter(model6.results, Phylum == PH[i], Term == "time:intB")
pv <- ifelse(pv[7] >= .001, sprintf("= %.3f", pv[7]), "< 0.001")
pL[[PH[i]]] <- ggplot(idat, aes(Week, PRED, color=Intervention, group=SubjectID))+
geom_line() +
#geom_jitter(aes(x=Week, y=Outcome),
# width=0.25, height = 0)+
scale_x_continuous(breaks=c(0,4,8,12))+
scale_y_continuous(limits=c(-5,10), breaks=seq(-5,10,2.5))+
labs(y="log(Abundance)", title=paste0(PH[i])) +
annotate("text", x=6, y=10, label=paste0("p ", pv))+
theme_classic() +
theme(legend.position = c(0.5,0.5))
}
# extract legend
leg <- get_legend(pL[[1]])
# some specializing for more space
pL[[1]] <- pL[[1]] + theme(legend.position = "none",
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
pL[[2]] <- pL[[2]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[3]] <- pL[[3]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[4]] <- pL[[4]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[5]] <- pL[[5]] + theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank())
pL[[6]] <- pL[[6]] + theme(legend.position = "none")
pL[[7]] <- pL[[7]] + theme(legend.position = "none",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())
pL[[8]] <- pL[[8]] + theme(legend.position = "none",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())
pL[[9]] <- pL[[9]] + theme(legend.position = "none",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())
# merge plot
p <- (pL[[1]] + pL[[2]] + pL[[3]] + pL[[4]] + pL[[5]] + pL[[6]] + pL[[7]] + pL[[8]] + pL[[9]] + leg) + plot_layout(ncol=5, nrow=2)
p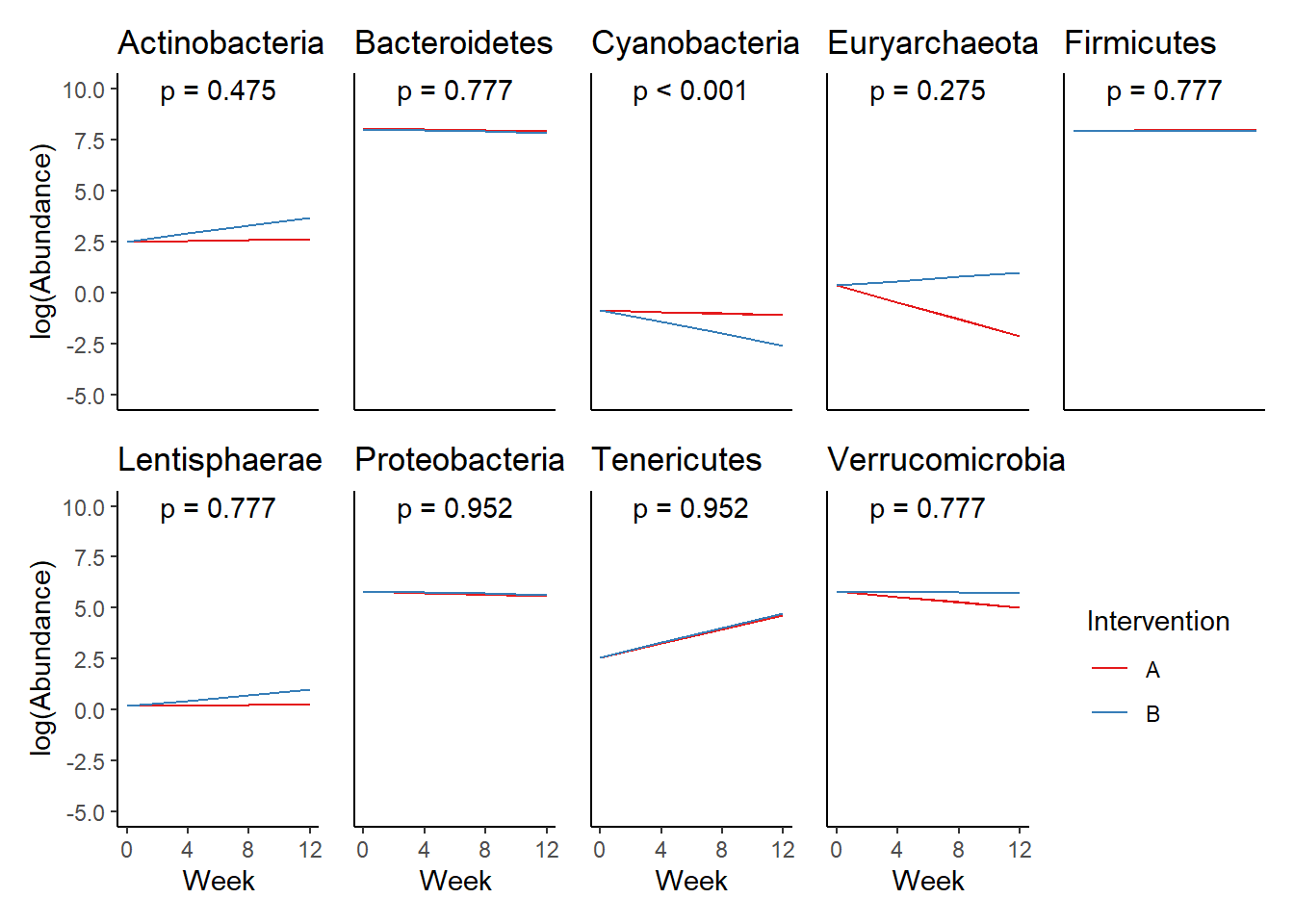
#ggsave("fig/figure5_glmm_phylum.pdf", p, units="in", width=10, height=4)
sessionInfo()R version 3.6.3 (2020-02-29)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 18362)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.0.0 microbiome_1.8.0 car_3.0-8 carData_3.0-4
[5] gvlma_1.0.0.3 patchwork_1.0.0 viridis_0.5.1 viridisLite_0.3.0
[9] gridExtra_2.3 xtable_1.8-4 kableExtra_1.1.0 plyr_1.8.6
[13] data.table_1.12.8 readxl_1.3.1 forcats_0.5.0 stringr_1.4.0
[17] dplyr_0.8.5 purrr_0.3.4 readr_1.3.1 tidyr_1.1.0
[21] tibble_3.0.1 ggplot2_3.3.0 tidyverse_1.3.0 lmerTest_3.1-2
[25] lme4_1.1-23 Matrix_1.2-18 vegan_2.5-6 lattice_0.20-38
[29] permute_0.9-5 phyloseq_1.30.0
loaded via a namespace (and not attached):
[1] Rtsne_0.15 minqa_1.2.4 colorspace_1.4-1
[4] rio_0.5.16 ellipsis_0.3.1 rprojroot_1.3-2
[7] XVector_0.26.0 fs_1.4.1 rstudioapi_0.11
[10] farver_2.0.3 fansi_0.4.1 lubridate_1.7.8
[13] xml2_1.3.2 codetools_0.2-16 splines_3.6.3
[16] knitr_1.28 ade4_1.7-15 jsonlite_1.6.1
[19] workflowr_1.6.2 nloptr_1.2.2.1 broom_0.5.6
[22] cluster_2.1.0 dbplyr_1.4.4 BiocManager_1.30.10
[25] compiler_3.6.3 httr_1.4.1 backports_1.1.7
[28] assertthat_0.2.1 cli_2.0.2 later_1.0.0
[31] htmltools_0.4.0 tools_3.6.3 igraph_1.2.5
[34] gtable_0.3.0 glue_1.4.1 reshape2_1.4.4
[37] Rcpp_1.0.4.6 Biobase_2.46.0 cellranger_1.1.0
[40] vctrs_0.3.0 Biostrings_2.54.0 multtest_2.42.0
[43] ape_5.3 nlme_3.1-144 iterators_1.0.12
[46] xfun_0.14 openxlsx_4.1.5 rvest_0.3.5
[49] lifecycle_0.2.0 statmod_1.4.34 zlibbioc_1.32.0
[52] MASS_7.3-51.5 scales_1.1.1 hms_0.5.3
[55] promises_1.1.0 parallel_3.6.3 biomformat_1.14.0
[58] rhdf5_2.30.1 RColorBrewer_1.1-2 curl_4.3
[61] yaml_2.2.1 stringi_1.4.6 highr_0.8
[64] S4Vectors_0.24.4 foreach_1.5.0 BiocGenerics_0.32.0
[67] zip_2.0.4 boot_1.3-24 rlang_0.4.6
[70] pkgconfig_2.0.3 evaluate_0.14 Rhdf5lib_1.8.0
[73] labeling_0.3 tidyselect_1.1.0 magrittr_1.5
[76] R6_2.4.1 IRanges_2.20.2 generics_0.0.2
[79] DBI_1.1.0 foreign_0.8-75 pillar_1.4.4
[82] haven_2.3.0 withr_2.2.0 mgcv_1.8-31
[85] abind_1.4-5 survival_3.1-8 modelr_0.1.8
[88] crayon_1.3.4 rmarkdown_2.1 grid_3.6.3
[91] blob_1.2.1 git2r_0.27.1 reprex_0.3.0
[94] digest_0.6.25 webshot_0.5.2 httpuv_1.5.2
[97] numDeriv_2016.8-1.1 stats4_3.6.3 munsell_0.5.0