Obesity Prevention
Obesity is a disease affecting more than one-third of our population, and is on the rise in countries with improving economies. We know that as people adopt Western lifestyles, both their diet and microbiome change dramatically, increasing obesity incidence. Dietary intake is one the most important factors shaping our resident microbial ecosystem, which also contributes to obesity development. We have been conducting studies to better understand the relationship between dietary intake, the microbiome, and weight gain. Currently, we are conducting a dietary fiber intervention among medical residents to study the effect on weight gain, body composition changes, appetite hormones.
Cancer Prevention
Colorectal cancer (CRC) is the second most lethal cancer in the U.S.. Alarmingly, incidence and mortality from CRC is on the rise among those under the age of 50. Several studies now highlight the microbiome as a key biomarker and contributor to colon cancer development. Diet is also key in preventing cancer, especially colon cancer. Unfortunately, very little focus has been placed on understanding the interaction between diet and the gut microbiome in cancer prevention and treatment response to cancer therapy. The standard treatment usually includes surgery and chemotherapy and now immunotherapy. These treatments, while demonstrating improvement in survival, often have severe side effects, such as severe diarrhea, that can be both transient and long-lasting. Studies from our research indicate that diet is likely a key factor in predicting treatment toxicity, and possible target for intervention. We are currently developing the tools necessary to arcuately capture dietary information during cancer treatment, as well as, microbiome data to better understand and identify the factors that predict cancer risk and response to therapy. Additionally, we previously identified a link between the lung microbiome and mutation in a tumor suppressor gene, TP53, and are studying this relationship in colon cancer.
Inflammation Prevention
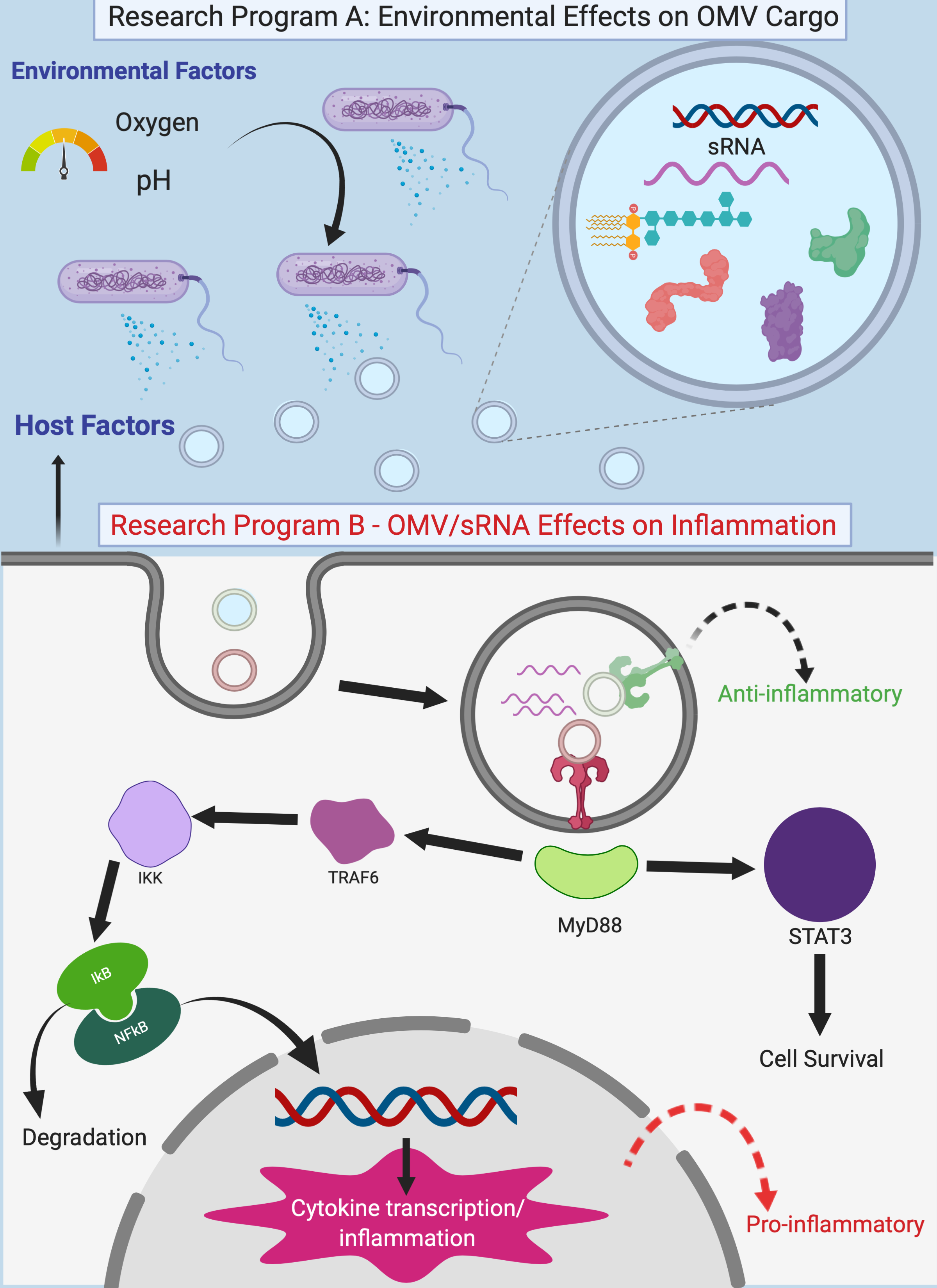
Microbiome-Host Communication
Chronic inflammatory diseases, which include both obesity and cancer, have been on the rise in Western countries for decades, yet clinical tools for prediction or curative treatments remain elusive. Bacteria in the colon play a key role in regulating the balance between acute and chronic inflammation, and understanding how to re-establish immune homeostasis is paramount in preventing chronic inflammatory disease development. The inflammatory response is governed, in part, through recognition of extracellular microbial RNA species by host cell receptors, including Toll-like Receptors (TLRs). Extracellular bacterial RNA is able to bind host TLRs and activate inflammatory signals, as well as provide tolerance to microbes. Recently, bacterial small RNA (sRNA) species were identified within outer membrane vesicles (OMVs) that are shed by bacteria. Small RNAs represent a critical trigger governing whether host cells respond with pro- or anti-inflammatory signals. It is unclear, however, how these sRNA species are parsed as cargo in OMVs, how sRNA species are transmitted from the bacteria to host intestinal epithelial cells, nor the mechanisms dictating host response to sRNA. We have evidence now showing that these OMV-derived small RNAs activate TLRs and the inflammatory pathways in epithelial cells. Defining the mechanisms by which bacteria use sRNA species to communicate with the host, and its role in controlling inflammation, holds promise for development of new strategies to control inflammation and re-establish intestinal homeostasis.
Microbiome Research Standardization
Human microbiome research has been exploding, but has outstripped standardization practices. Our lab has been leading the charge to improve access to improved protocols in human microbiome studies. Standardization in microbiome analyses is fundamental to reliably measure the human microbiome. Variability in results can stem from all the steps in the human microbiome study process including sample collection, DNA extraction, sequencing, and bioinformatics. Of these, DNA extraction was identified by the MicroBiome Quality Control project (MBQC), the International Human Microbiome Standards (IHMS) group, and others as contributing a majority of experimental variability. Together with other labs, we set forth a list of best practices for laboratories considering human microbiome studies, especially those working with low biomass samples (e.g. fresh-frozen tissues). Our goal is to continue to improve protocols and to help other researchers produce high-quality and reproducible microbiome research.